+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TLR3-mAb12-poly(I:C) | |||||||||
 Map data Map data | TLR3-mAb12-poly(I:C) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 13.2 Å | |||||||||
 Authors Authors | Lim CS / Jang YH / Lee GY / Han GM / Lee JO | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand. Authors: Chan Seok Lim / Yoon Ha Jang / Ga Young Lee / Gu Min Han / Hye Jin Jeong / Ji Won Kim / Jie-Oh Lee /  Abstract: Toll-like Receptor 3 (TLR3) initiates a potent anti-viral immune response by binding to double-stranded RNA ligands. Previous crystallographic studies showed that TLR3 forms a homodimer when bound to ...Toll-like Receptor 3 (TLR3) initiates a potent anti-viral immune response by binding to double-stranded RNA ligands. Previous crystallographic studies showed that TLR3 forms a homodimer when bound to a 46-base pair RNA ligand. However, this short RNA fails to initiate a robust immune response. To obtain structural insights into the length dependency of TLR3 ligands, we determine the cryo-electron microscopy structure of full-length TLR3 in a complex with a synthetic RNA ligand with an average length of ~400 base pairs. In the structure, the dimeric TLR3 units are clustered along the double-stranded RNA helix in a highly organized and cooperative fashion with a uniform inter-dimer spacing of 103 angstroms. The intracellular and transmembrane domains are dispensable for the clustering because their deletion does not interfere with the cluster formation. Our structural observation suggests that ligand-induced clustering of TLR3 dimers triggers the ordered assembly of intracellular signaling adaptors and initiates a robust innate immune response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34367.map.gz emd_34367.map.gz | 229.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34367-v30.xml emd-34367-v30.xml emd-34367.xml emd-34367.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34367.png emd_34367.png | 29.1 KB | ||
| Others |  emd_34367_half_map_1.map.gz emd_34367_half_map_1.map.gz emd_34367_half_map_2.map.gz emd_34367_half_map_2.map.gz | 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34367 http://ftp.pdbj.org/pub/emdb/structures/EMD-34367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34367 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34367.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34367.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TLR3-mAb12-poly(I:C) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
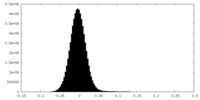
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: low-pass filtered (> 7 angstrom)
| File | emd_34367_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | low-pass filtered (> 7 angstrom) | ||||||||||||
| Projections & Slices |
| ||||||||||||
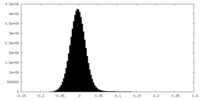
| Density Histograms |
-Half map: low-pass filtered (> 7 angstrom)
| File | emd_34367_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | low-pass filtered (> 7 angstrom) | ||||||||||||
| Projections & Slices |
| ||||||||||||
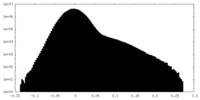
| Density Histograms |
- Sample components
Sample components
-Entire : TLR3-mAb12-poly(I:C)
| Entire | Name: TLR3-mAb12-poly(I:C) |
|---|---|
| Components |
|
-Supramolecule #1: TLR3-mAb12-poly(I:C)
| Supramolecule | Name: TLR3-mAb12-poly(I:C) / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 / Details: Fab form of mAb12 bound to TLR3-poly(I:C) complex |
|---|
-Supramolecule #2: TLR3
| Supramolecule | Name: TLR3 / type: complex / Chimera: Yes / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: mAb12
| Supramolecule | Name: mAb12 / type: complex / Chimera: Yes / ID: 3 / Parent: 1 / Macromolecule list: #2 / Details: Fab form |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: poly(I:C)
| Supramolecule | Name: poly(I:C) / type: complex / Chimera: Yes / ID: 4 / Parent: 1 / Macromolecule list: #3-#4 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.26 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 5.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 13.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.2) / Number images used: 6864 |
|---|---|
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)