[English] 日本語
 Yorodumi
Yorodumi- EMDB-34218: Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR / RNase / RNA BINDING PROTEIN-RNA COMPLEX | |||||||||
| Function / homology | CHAT domain / CHAT domain / : / CRISPR type III-associated protein / RAMP superfamily / defense response to virus / CRISPR-associated RAMP family protein / CHAT domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  Desulfonema ishimotonii (bacteria) Desulfonema ishimotonii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.84 Å | |||||||||
 Authors Authors | Kato K / Okazaki S / Ishikawa J / Isayama Y / Nishizawa T / Nishimasu H | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease. Authors: Kazuki Kato / Sae Okazaki / Cian Schmitt-Ulms / Kaiyi Jiang / Wenyuan Zhou / Junichiro Ishikawa / Yukari Isayama / Shungo Adachi / Tomohiro Nishizawa / Kira S Makarova / Eugene V Koonin / ...Authors: Kazuki Kato / Sae Okazaki / Cian Schmitt-Ulms / Kaiyi Jiang / Wenyuan Zhou / Junichiro Ishikawa / Yukari Isayama / Shungo Adachi / Tomohiro Nishizawa / Kira S Makarova / Eugene V Koonin / Omar O Abudayyeh / Jonathan S Gootenberg / Hiroshi Nishimasu /   Abstract: The type III-E CRISPR-Cas7-11 effector binds a CRISPR RNA (crRNA) and the putative protease Csx29 and catalyzes crRNA-guided RNA cleavage. We report cryo-electron microscopy structures of the Cas7-11- ...The type III-E CRISPR-Cas7-11 effector binds a CRISPR RNA (crRNA) and the putative protease Csx29 and catalyzes crRNA-guided RNA cleavage. We report cryo-electron microscopy structures of the Cas7-11-crRNA-Csx29 complex with and without target RNA (tgRNA), and demonstrate that tgRNA binding induces conformational changes in Csx29. Biochemical experiments revealed tgRNA-dependent cleavage of the accessory protein Csx30 by Csx29. Reconstitution of the system in bacteria showed that Csx30 cleavage yields toxic protein fragments that cause growth arrest, which is regulated by Csx31. Csx30 binds Csx31 and the associated sigma factor RpoE (RNA polymerase, extracytoplasmic E), suggesting that Csx30-mediated RpoE inhibition modulates the cellular response to infection. We engineered the Cas7-11-Csx29-Csx30 system for programmable RNA sensing in mammalian cells. Overall, the Cas7-11-Csx29 effector is an RNA-dependent nuclease-protease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34218.map.gz emd_34218.map.gz | 168.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34218-v30.xml emd-34218-v30.xml emd-34218.xml emd-34218.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34218.png emd_34218.png | 59.4 KB | ||
| Masks |  emd_34218_msk_1.map emd_34218_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-34218.cif.gz emd-34218.cif.gz | 7.3 KB | ||
| Others |  emd_34218_half_map_1.map.gz emd_34218_half_map_1.map.gz emd_34218_half_map_2.map.gz emd_34218_half_map_2.map.gz | 165.2 MB 165.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34218 http://ftp.pdbj.org/pub/emdb/structures/EMD-34218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34218 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34218 | HTTPS FTP |
-Related structure data
| Related structure data |  8gs2MC  7y9xC  7y9yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34218.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34218.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
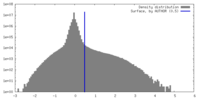
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34218_msk_1.map emd_34218_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
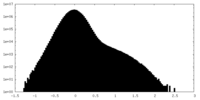
| Density Histograms |
-Half map: #1
| File | emd_34218_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
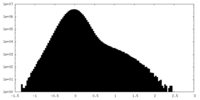
| Density Histograms |
-Half map: #2
| File | emd_34218_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Cas7-11 in complex with a crRNA and its target RNA
+Supramolecule #1: Cas7-11 in complex with a crRNA and its target RNA
+Supramolecule #2: crRNA
+Supramolecule #3: target RNA with 3'-tag
+Supramolecule #4: Cas7-11, Csx29
+Macromolecule #1: crRNA
+Macromolecule #2: target RNA with non-matching PFS
+Macromolecule #3: CRISPR-associated RAMP family protein
+Macromolecule #4: CHAT domain-containing protein
+Macromolecule #5: CYTIDINE-5'-MONOPHOSPHATE
+Macromolecule #6: ADENOSINE MONOPHOSPHATE
+Macromolecule #7: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 76.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.84 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 71543 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)