+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FTY720p-bound human SPNS2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of eye pigmentation / regulation of humoral immune response / regulation of T cell migration / sphingolipid transporter activity / lymphocyte migration / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / sphingosine-1-phosphate receptor signaling pathway / lipid transport / T cell homeostasis ...regulation of eye pigmentation / regulation of humoral immune response / regulation of T cell migration / sphingolipid transporter activity / lymphocyte migration / sphingolipid biosynthetic process / Sphingolipid de novo biosynthesis / sphingosine-1-phosphate receptor signaling pathway / lipid transport / T cell homeostasis / B cell homeostasis / transmembrane transporter activity / lymph node development / sensory perception of sound / bone development / endosome membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
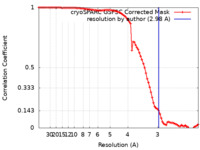
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | He Y / Duan Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Structural basis of Sphingosine-1-phosphate transport via human SPNS2. Authors: Yaning Duan / Nancy C P Leong / Jing Zhao / Yu Zhang / Dat T Nguyen / Hoa T T Ha / Na Wang / Ruixue Xia / Zhenmei Xu / Zhengxiong Ma / Yu Qian / Han Yin / Xinyan Zhu / Anqi Zhang / Changyou ...Authors: Yaning Duan / Nancy C P Leong / Jing Zhao / Yu Zhang / Dat T Nguyen / Hoa T T Ha / Na Wang / Ruixue Xia / Zhenmei Xu / Zhengxiong Ma / Yu Qian / Han Yin / Xinyan Zhu / Anqi Zhang / Changyou Guo / Yu Xia / Long N Nguyen / Yuanzheng He /   | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34104.map.gz emd_34104.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34104-v30.xml emd-34104-v30.xml emd-34104.xml emd-34104.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34104_fsc.xml emd_34104_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_34104.png emd_34104.png | 64.8 KB | ||
| Filedesc metadata |  emd-34104.cif.gz emd-34104.cif.gz | 6.2 KB | ||
| Others |  emd_34104_half_map_1.map.gz emd_34104_half_map_1.map.gz emd_34104_half_map_2.map.gz emd_34104_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34104 http://ftp.pdbj.org/pub/emdb/structures/EMD-34104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34104 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34104 | HTTPS FTP |
-Validation report
| Summary document |  emd_34104_validation.pdf.gz emd_34104_validation.pdf.gz | 814.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34104_full_validation.pdf.gz emd_34104_full_validation.pdf.gz | 814.4 KB | Display | |
| Data in XML |  emd_34104_validation.xml.gz emd_34104_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_34104_validation.cif.gz emd_34104_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34104 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34104 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34104 | HTTPS FTP |
-Related structure data
| Related structure data |  7yudMC  7yubC  7yufC  8kaeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34104.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34104.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
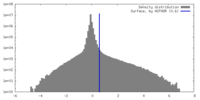
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34104_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
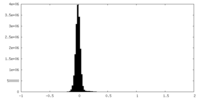
| Density Histograms |
-Half map: #1
| File | emd_34104_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
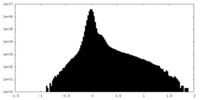
| Density Histograms |
- Sample components
Sample components
-Entire : SPNS2 nanobody complex
| Entire | Name: SPNS2 nanobody complex |
|---|---|
| Components |
|
-Supramolecule #1: SPNS2 nanobody complex
| Supramolecule | Name: SPNS2 nanobody complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: NbFab-H-chain
| Macromolecule | Name: NbFab-H-chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 26.644539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEISEVQLVE SGGGLVQPGG SLRLSCAASG FNFSYYSIHW VRQAPGKGLE WVAYISSSSS YTSYADSVKG RFTISADTSK NTAYLQMNS LRAEDTAVYY CARGYQYWQY HASWYWNGGL DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP ...String: MEISEVQLVE SGGGLVQPGG SLRLSCAASG FNFSYYSIHW VRQAPGKGLE WVAYISSSSS YTSYADSVKG RFTISADTSK NTAYLQMNS LRAEDTAVYY CARGYQYWQY HASWYWNGGL DYWGQGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC L VKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDKT HT HHHHHH |
-Macromolecule #2: NbFab L-chain
| Macromolecule | Name: NbFab L-chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.38998 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDIQMTQSP SSLSASVGDR VTITCRASQS VSSAVAWYQQ KPGKAPKLLI YSASSLYSGV PSRFSGSRSG TDFTLTISSL QPEDFATYY CQQSSSSLIT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ ...String: MSDIQMTQSP SSLSASVGDR VTITCRASQS VSSAVAWYQQ KPGKAPKLLI YSASSLYSGV PSRFSGSRSG TDFTLTISSL QPEDFATYY CQQSSSSLIT FGQGTKVEIK RTVAAPSVFI FPPSDSQLKS GTASVVCLLN NFYPREAKVQ WKVDNALQSG N SQESVTEQ DSKDSTYSLS STLTLSKADY EKHKVYACEV THQGLSSPVT KSFNRGEC |
-Macromolecule #3: Tc-Nb8
| Macromolecule | Name: Tc-Nb8 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.001732 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QRQLVESGGG LVQPGGSLRL SCAAPGSRRF DYYTLGWFRQ APGKEREGVS CISTVGGITN YADSVKGRFI ISRDNAKSTV YLQMNSLEP EDTAVYYCAA GREMCAPMML GDYYDMDYWG KGTPVTVSS |
-Macromolecule #4: Sphingosine-1-phosphate transporter SPNS2
| Macromolecule | Name: Sphingosine-1-phosphate transporter SPNS2 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.096289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMCLECASAA AGGAEEEEAD AERRRRRRGA QRGAGGSGCC GARGAGGAGV SAAGDEVQTL SGSVRRAPTG PPGTPGTPGC AATAKGPGA QQPKPASLGR GRGAAAAILS LGNVLNYLDR YTVAGVLLDI QQHFGVKDRG AGLLQSVFIC SFMVAAPIFG Y LGDRFNRK ...String: MMCLECASAA AGGAEEEEAD AERRRRRRGA QRGAGGSGCC GARGAGGAGV SAAGDEVQTL SGSVRRAPTG PPGTPGTPGC AATAKGPGA QQPKPASLGR GRGAAAAILS LGNVLNYLDR YTVAGVLLDI QQHFGVKDRG AGLLQSVFIC SFMVAAPIFG Y LGDRFNRK VILSCGIFFW SAVTFSSSFI PQQYFWLLVL SRGLVGIGEA SYSTIAPTII GDLFTKNTRT LMLSVFYFAI PL GSGLGYI TGSSVKQAAG DWHWALRVSP VLGMITGTLI LILVPATKRG HADQLGDQLK ARTSWLRDMK ALIRNRSYVF SSL ATSAVS FATGALGMWI PLYLHRAQVV QKTAETCNSP PCGAKDSLIF GAITCFTGFL GVVTGAGATR WCRLKTQRAD PLVC AVGML GSAIFICLIF VAAKSSIVGA YICIFVGETL LFSNWAITAD ILMYVVIPTR RATAVALQSF TSHLLGDAGS PYLIG FISD LIRQSTKDSP LWEFLSLGYA LMLCPFVVVL GGMFFLATAL FFVSDRARAE QQVNQLAMPP ASVKV UniProtKB: Sphingosine-1-phosphate transporter SPNS2 |
-Macromolecule #5: anti-Fab nanobody
| Macromolecule | Name: anti-Fab nanobody / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.76411 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSQVQLQES GGGLVQPGGS LRLSCAASGR TISRYAMSWF RQAPGKEREF VAVARRSGDG AFYADSVQGR FTVSRDDAKN TVYLQMNSL KPEDTAVYYC AIDSDTFYSG SYDYWGQGTQ VTVSSLE |
-Macromolecule #6: (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene...
| Macromolecule | Name: (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol type: ligand / ID: 6 / Number of copies: 1 / Formula: J89 |
|---|---|
| Molecular weight | Theoretical: 387.451 Da |
| Chemical component information |  ChemComp-J89: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)