+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of baculovirus LEF-3 in complex with ssDNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ssDNA / SSB / Baculovirus / DNA BINDING PROTEIN | |||||||||
| Function / homology | Nucleopolyhedrovirus late expression factor 3 / Nucleopolyhedrovirus late expression factor 3 (LEF-3) / regulation of DNA-templated transcription / DNA binding / Lef3 Function and homology information Function and homology information | |||||||||
| Biological species |  Helicoverpa armigera nucleopolyhedrovirus / synthetic construct (others) Helicoverpa armigera nucleopolyhedrovirus / synthetic construct (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Yin J / Fu Y / Rao G / Li Z / Cao S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Structural transitions during the cooperative assembly of baculovirus single-stranded DNA-binding protein on ssDNA. Authors: Jiayi Yin / Yan Fu / Guibo Rao / Zhiqiang Li / Kexing Tian / Tingting Chong / Kai Kuang / Manli Wang / Zhihong Hu / Sheng Cao /  Abstract: Single-stranded DNA-binding proteins (SSBs) interact with single-stranded DNA (ssDNA) to form filamentous structures with various degrees of cooperativity, as a result of intermolecular interactions ...Single-stranded DNA-binding proteins (SSBs) interact with single-stranded DNA (ssDNA) to form filamentous structures with various degrees of cooperativity, as a result of intermolecular interactions between neighboring SSB subunits on ssDNA. However, it is still challenging to perform structural studies on SSB-ssDNA filaments at high resolution using the most studied SSB models, largely due to the intrinsic flexibility of these nucleoprotein complexes. In this study, HaLEF-3, an SSB protein from Helicoverpa armigera nucleopolyhedrovirus, was used for in vitro assembly of SSB-ssDNA filaments, which were structurally studied at atomic resolution using cryo-electron microscopy. Combined with the crystal structure of ssDNA-free HaLEF-3 octamers, our results revealed that the three-dimensional rearrangement of HaLEF-3 induced by an internal hinge-bending movement is essential for the formation of helical SSB-ssDNA complexes, while the contacting interface between adjacent HaLEF-3 subunits remains basically intact. We proposed a local cooperative SSB-ssDNA binding model, in which, triggered by exposure to oligonucleotides, HaLEF-3 molecules undergo ring-to-helix transition to initiate continuous SSB-SSB interactions along ssDNA. Unique structural features revealed by the assembly of HaLEF-3 on ssDNA suggest that HaLEF-3 may represent a new class of SSB. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34010.map.gz emd_34010.map.gz | 49.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34010-v30.xml emd-34010-v30.xml emd-34010.xml emd-34010.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34010.png emd_34010.png | 159.3 KB | ||
| Filedesc metadata |  emd-34010.cif.gz emd-34010.cif.gz | 5.5 KB | ||
| Others |  emd_34010_half_map_1.map.gz emd_34010_half_map_1.map.gz emd_34010_half_map_2.map.gz emd_34010_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34010 http://ftp.pdbj.org/pub/emdb/structures/EMD-34010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34010 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34010 | HTTPS FTP |
-Related structure data
| Related structure data |  7ypoMC  7ynyC  7ypqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34010.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34010.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
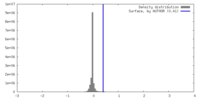
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34010_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
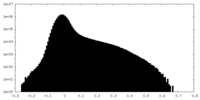
| Projections & Slices |
| ||||||||||||
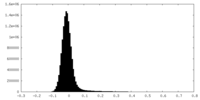
| Density Histograms |
-Half map: #1
| File | emd_34010_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
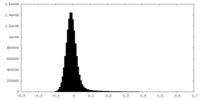
| Density Histograms |
- Sample components
Sample components
-Entire : Baculovirus LEF-3 in complex with ssDNA
| Entire | Name: Baculovirus LEF-3 in complex with ssDNA |
|---|---|
| Components |
|
-Supramolecule #1: Baculovirus LEF-3 in complex with ssDNA
| Supramolecule | Name: Baculovirus LEF-3 in complex with ssDNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: LEF-3
| Supramolecule | Name: LEF-3 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Helicoverpa armigera nucleopolyhedrovirus Helicoverpa armigera nucleopolyhedrovirus |
-Supramolecule #3: ssDNA
| Supramolecule | Name: ssDNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Lef3
| Macromolecule | Name: Lef3 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Helicoverpa armigera nucleopolyhedrovirus Helicoverpa armigera nucleopolyhedrovirus |
| Molecular weight | Theoretical: 47.637707 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMSNMDI SPVKQLIDIE NDDAMNTPEK GMKRPLMRTM SSVEEPQAKM AKLRTLNVK GQLLTKTTMS INNEDYYLFK FLVNNKSIDY YGTQTQFFSL INNKTYELVL QYSRKKLLIK SYEQCEDEDL L MTVCKSVT ...String: MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSMSNMDI SPVKQLIDIE NDDAMNTPEK GMKRPLMRTM SSVEEPQAKM AKLRTLNVK GQLLTKTTMS INNEDYYLFK FLVNNKSIDY YGTQTQFFSL INNKTYELVL QYSRKKLLIK SYEQCEDEDL L MTVCKSVT FQEFCANEIK SLLAKFLYGF KIYGSSNVYK LVFVILLEDN NGTINGVQVE MMSDFKRLSG AFKNHVIENE ND LFDCMYK SEEKYFNLYR IKCNHNANNY KSLSLSSNSQ LERLETDDSM FEYEFQYDYT VNISRSNKII QKHRVTGNFT SER NIYQNS DRFVISYDTA NEKIKTSIYN RMENAESKTD YDTSITLKDV TLSQLNSLIE SNLVQVDVYL VTDPNNVKNN VIAG ITKIE IDGTYEPL UniProtKB: Lef3 |
-Macromolecule #2: DNA (28-MER)
| Macromolecule | Name: DNA (28-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.724834 KDa |
| Sequence | String: (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) (DA)(DA)(DA)(DA)(DA)(DA)(DA)(DA) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.3 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 15.105 Å Applied symmetry - Helical parameters - Δ&Phi: -108.281 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 530580 |
|---|---|
| Startup model | Type of model: NONE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)