[English] 日本語
 Yorodumi
Yorodumi- EMDB-33985: Complex structure of Neuropeptide Y Y2 receptor in complex with N... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
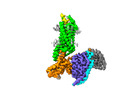
| Title | Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi | |||||||||
 Map data Map data | Composite map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of nervous system process / regulation of nerve growth factor production / peptide YY receptor activity / short-day photoperiodism / negative regulation of acute inflammatory response to antigenic stimulus / positive regulation of circadian sleep/wake cycle, non-REM sleep / positive regulation of peptide secretion / neuropeptide Y receptor activity / monocyte activation / positive regulation of dopamine metabolic process ...negative regulation of nervous system process / regulation of nerve growth factor production / peptide YY receptor activity / short-day photoperiodism / negative regulation of acute inflammatory response to antigenic stimulus / positive regulation of circadian sleep/wake cycle, non-REM sleep / positive regulation of peptide secretion / neuropeptide Y receptor activity / monocyte activation / positive regulation of dopamine metabolic process / positive regulation of nitric oxide metabolic process / positive regulation of appetite / neuropeptide Y receptor binding / intestinal epithelial cell differentiation / synaptic signaling via neuropeptide / positive regulation of eating behavior / negative regulation of secretion / cardiac left ventricle morphogenesis / adult feeding behavior / positive regulation of smooth muscle contraction / positive regulation of dopamine secretion / regulation of presynaptic cytosolic calcium ion concentration / negative regulation of excitatory postsynaptic potential / neuropeptide hormone activity / feeding behavior / negative regulation of synaptic transmission, glutamatergic / non-motile cilium / negative regulation of cAMP/PKA signal transduction / positive regulation of cell-substrate adhesion / negative regulation of feeding behavior / regulation of synaptic vesicle exocytosis / central nervous system neuron development / outflow tract morphogenesis / nitric oxide mediated signal transduction / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / neuropeptide signaling pathway / behavioral fear response / FOXO-mediated transcription of oxidative stress, metabolic and neuronal genes / neuronal dense core vesicle / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / negative regulation of blood pressure / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Peptide ligand-binding receptors / calcium channel regulator activity / Regulation of insulin secretion / locomotory behavior / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor activity / G protein-coupled receptor binding / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / cerebral cortex development / GABA-ergic synapse / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / terminal bouton / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through CDC42 / Glucagon signaling in metabolic regulation / neuron projection development / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / GDP binding / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / cellular response to prostaglandin E stimulus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.11 Å | |||||||||
 Authors Authors | Kang H / Park C / Kim J / Choi H-J | |||||||||
| Funding support |  Korea, Republic Of, 2 items Korea, Republic Of, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling. Authors: Hyunook Kang / Chaehee Park / Yeol Kyo Choi / Jungnam Bae / Sohee Kwon / Jinuk Kim / Chulwon Choi / Chaok Seok / Wonpil Im / Hee-Jung Choi /   Abstract: Neuropeptide Y (NPY) and its receptors are expressed in various human tissues including the brain where they regulate appetite and emotion. Upon NPY stimulation, the neuropeptide Y1 and Y2 receptors ...Neuropeptide Y (NPY) and its receptors are expressed in various human tissues including the brain where they regulate appetite and emotion. Upon NPY stimulation, the neuropeptide Y1 and Y2 receptors (YR and YR, respectively) activate G signaling, but their physiological responses to food intake are different. In addition, deletion of the two N-terminal amino acids of peptide YY (PYY(3-36)), the endogenous form found in circulation, can stimulate YR but not YR, suggesting that YR and YR may have distinct ligand-binding modes. Here, we report the cryo-electron microscopy structures of the PYY(3-36)‒YR‒G and NPY‒YR‒G complexes. Using cell-based assays, molecular dynamics simulations, and structural analysis, we revealed the molecular basis of the exclusive binding of PYY(3-36) to YR. Furthermore, we demonstrated that YR favors G protein signaling over β-arrestin signaling upon activation, whereas YR does not show a preference between these two pathways. #1:  Journal: Acta Crystallogr D Struct Biol / Year: 2018 Journal: Acta Crystallogr D Struct Biol / Year: 2018Title: Real-space refinement in PHENIX for cryo-EM and crystallography. Authors: Afonine PV / Poon BK / Read RJ / Sobolev OV / Terwilliger TC / Urzhumtsev A / Adams PD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33985.map.gz emd_33985.map.gz | 229.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33985-v30.xml emd-33985-v30.xml emd-33985.xml emd-33985.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33985.png emd_33985.png | 43.1 KB | ||
| Others |  emd_33985_additional_1.map.gz emd_33985_additional_1.map.gz emd_33985_additional_2.map.gz emd_33985_additional_2.map.gz | 230 MB 230.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33985 http://ftp.pdbj.org/pub/emdb/structures/EMD-33985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33985 | HTTPS FTP |
-Validation report
| Summary document |  emd_33985_validation.pdf.gz emd_33985_validation.pdf.gz | 429.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33985_full_validation.pdf.gz emd_33985_full_validation.pdf.gz | 429.3 KB | Display | |
| Data in XML |  emd_33985_validation.xml.gz emd_33985_validation.xml.gz | 7.1 KB | Display | |
| Data in CIF |  emd_33985_validation.cif.gz emd_33985_validation.cif.gz | 8.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33985 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33985 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33985 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33985 | HTTPS FTP |
-Related structure data
| Related structure data |  7yooMC  7yonC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33985.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33985.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8415 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Local refined map of NPY-Y2R
| File | emd_33985_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refined map of NPY-Y2R | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Local refined map of G-protein complex
| File | emd_33985_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refined map of G-protein complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Complex structure of NPY-Y2R-Gi-scFv16
+Supramolecule #1: Complex structure of NPY-Y2R-Gi-scFv16
+Supramolecule #2: Guanine nucleotide-binding protein
+Supramolecule #3: scFv16
+Supramolecule #4: NPY
+Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
+Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
+Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
+Macromolecule #4: Neuropeptide Y
+Macromolecule #5: Neuropeptide Y receptor type 2
+Macromolecule #6: single-chain antibody Fv fragment (scFv16)
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.25 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)
































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)

