+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of CasPi with guide RNA and target DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CasPi complex / RNA BINDING PROTEIN / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology | Transposase Function and homology information Function and homology information | |||||||||
| Biological species |  Armatimonadetes bacterium (bacteria) / Armatimonadetes bacterium (bacteria) /  Armatimonadota bacterium (bacteria) Armatimonadota bacterium (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.36 Å | |||||||||
 Authors Authors | Li CP / Wang J / Liu JJ / Zhang S | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: The compact Casπ (Cas12l) 'bracelet' provides a unique structural platform for DNA manipulation. Authors: Ao Sun / Cheng-Ping Li / Zhihang Chen / Shouyue Zhang / Dan-Yuan Li / Yun Yang / Long-Qi Li / Yuqian Zhao / Kaichen Wang / Zhaofu Li / Jinxia Liu / Sitong Liu / Jia Wang / Jun-Jie Gogo Liu /  Abstract: CRISPR-Cas modules serve as the adaptive nucleic acid immune systems for prokaryotes, and provide versatile tools for nucleic acid manipulation in various organisms. Here, we discovered a new ...CRISPR-Cas modules serve as the adaptive nucleic acid immune systems for prokaryotes, and provide versatile tools for nucleic acid manipulation in various organisms. Here, we discovered a new miniature type V system, CRISPR-Casπ (Cas12l) (~860 aa), from the environmental metagenome. Complexed with a large guide RNA (~170 nt) comprising the tracrRNA and crRNA, Casπ (Cas12l) recognizes a unique 5' C-rich PAM for DNA cleavage under a broad range of biochemical conditions, and generates gene editing in mammalian cells. Cryo-EM study reveals a 'bracelet' architecture of Casπ effector encircling the DNA target at 3.4 Å resolution, substantially different from the canonical 'two-lobe' architectures of Cas12 and Cas9 nucleases. The large guide RNA serves as a 'two-arm' scaffold for effector assembly. Our study expands the knowledge of DNA targeting mechanisms by CRISPR effectors, and offers an efficient but compact platform for DNA manipulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33983.map.gz emd_33983.map.gz | 49.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33983-v30.xml emd-33983-v30.xml emd-33983.xml emd-33983.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33983.png emd_33983.png | 106.5 KB | ||
| Filedesc metadata |  emd-33983.cif.gz emd-33983.cif.gz | 6.9 KB | ||
| Others |  emd_33983_half_map_1.map.gz emd_33983_half_map_1.map.gz emd_33983_half_map_2.map.gz emd_33983_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33983 http://ftp.pdbj.org/pub/emdb/structures/EMD-33983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33983 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33983 | HTTPS FTP |
-Related structure data
| Related structure data |  7yojMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33983.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33983.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.856 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33983_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
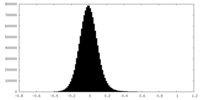
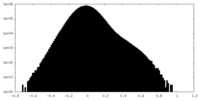
| Density Histograms |
-Half map: #1
| File | emd_33983_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
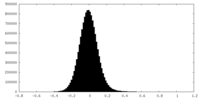
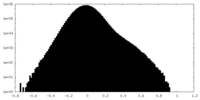
| Density Histograms |
- Sample components
Sample components
-Entire : CasPi
| Entire | Name: CasPi |
|---|---|
| Components |
|
-Supramolecule #1: CasPi
| Supramolecule | Name: CasPi / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Armatimonadetes bacterium (bacteria) Armatimonadetes bacterium (bacteria) |
| Molecular weight | Theoretical: 177.28 KDa |
-Macromolecule #1: CasPi
| Macromolecule | Name: CasPi / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Armatimonadota bacterium (bacteria) Armatimonadota bacterium (bacteria) |
| Molecular weight | Theoretical: 98.731234 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKATKEVKS KRVEALRQVA YQRLERLERK AQKIGAHLRK PGKAADLQSL HYLLHKVEVE YHDIARNLEK DPTWTPKPKM RREKRAIVP ESGPAAPLPT TAKGEPGRPA NRHIPPPVPL DSARIPEDQQ SMGQGSGGRS WCSAPFVEVK LPPTQWSNVR E KLLKFRIE ...String: MAKATKEVKS KRVEALRQVA YQRLERLERK AQKIGAHLRK PGKAADLQSL HYLLHKVEVE YHDIARNLEK DPTWTPKPKM RREKRAIVP ESGPAAPLPT TAKGEPGRPA NRHIPPPVPL DSARIPEDQQ SMGQGSGGRS WCSAPFVEVK LPPTQWSNVR E KLLKFRIE DDADIVRRWA EAKFGSIETA RDGLRASAEI GTSPDVWRSF ISRAISNGKK DFEPLLSLDD DELTADATAE RV VRRWHQI DWVGRMLDSI LETVPSGVSK DTFRSRVESR LKTFHSSVNS FELKKRKDGT VERKRKHTNP QFPYLSPSAV SID PDVVTM EAVELLQMQP EERFAKDPND ANGRMRLRVL QAELGKARRE ALGRRGEKAP PWSGRKVFRG TTTRKREACL VWDK EAQAD GLYFALVMSG GPKIDDKRFV YMDGQPLQSD WQLHNGVAGK AKSCRAMPLI LKHDFLRWYH RHIKNHDVNA PLEKR CVHT TTQFVFVEPD EKKGLQPRLF IRPVFKFYDP VYEVPDSHSI DKKPDCRYLI GIARGVNYPY RAAVYDCETN SIIADK FVD GRKADWERIR NELAYHQRRR DLLRNSRASS AAIQREIRAI ARIRKRERGL NKVETVESIA RLVDWAEENL GKCNYCF VL ADLSSNLNLG RNNRVKHIAA IKEALINQMR KRGYRFKKSG KVDGVREESA WYTSAVAPSG WWAKKEEVDG AWKADKTR P LARKIGSYYC CEEIDGLHLR GVLKGLGRAK RLVLQSDDPS APTRRRGFGS ELFWDPYCTE LCGHAFPQGV VLDADFIGA FNIALRPLVR EELGKKAKAV DLADRHQTLN PTVALRCGVT AYEFVEVGGD PRGGLRKILL NPAEAVI UniProtKB: Transposase |
-Macromolecule #2: DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*GP*GP*AP*TP*GP*CP*CP*CP*AP*G)-3') / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Armatimonadetes bacterium (bacteria) Armatimonadetes bacterium (bacteria) |
| Molecular weight | Theoretical: 3.688405 KDa |
| Sequence | String: (DC)(DG)(DG)(DG)(DA)(DT)(DG)(DC)(DC)(DC) (DA)(DG) |
-Macromolecule #3: DNA (30-MER)
| Macromolecule | Name: DNA (30-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Armatimonadetes bacterium (bacteria) Armatimonadetes bacterium (bacteria) |
| Molecular weight | Theoretical: 9.192878 KDa |
| Sequence | String: (DG)(DT)(DT)(DC)(DA)(DC)(DC)(DA)(DG)(DG) (DG)(DT)(DG)(DT)(DC)(DG)(DC)(DC)(DC)(DT) (DG)(DG)(DG)(DC)(DA)(DT)(DC)(DC)(DC) (DG) |
-Macromolecule #4: RNA (174-MER)
| Macromolecule | Name: RNA (174-MER) / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Armatimonadetes bacterium (bacteria) Armatimonadetes bacterium (bacteria) |
| Molecular weight | Theoretical: 56.411703 KDa |
| Sequence | String: GUCUGCCGAA GACGCCGCAC GGAGCCUGGG CCGGAAUCGU AGAUCGAACG CGGCAUCGAA GCCCUGCAGC CCUUCGGGGC CAAGGCGGC GCAGCAAGCC UCUUUCAGGC GGCAGAGUCC UUUAGAGUGU GAGAGACACU CUAAAGGAAU GAAAGAGGGC G ACACCCUG GUGAAC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing #1
Image processing #1
+ Image processing #2
Image processing #2
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE |
|---|---|
| Output model |  PDB-7yoj: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)