[English] 日本語
 Yorodumi
Yorodumi- EMDB-33939: Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of recombinant RyR2 (Ca2+ dataset, class 2, open state) | |||||||||||||||||||||
 Map data Map data | Structure of recombinant RyR2 (Ca2 dataset, class 2, open state) | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationestablishment of protein localization to endoplasmic reticulum / type B pancreatic cell apoptotic process / Purkinje myocyte to ventricular cardiac muscle cell signaling / regulation of atrial cardiac muscle cell action potential / left ventricular cardiac muscle tissue morphogenesis / suramin binding / regulation of AV node cell action potential / regulation of SA node cell action potential / Stimuli-sensing channels / regulation of ventricular cardiac muscle cell action potential ...establishment of protein localization to endoplasmic reticulum / type B pancreatic cell apoptotic process / Purkinje myocyte to ventricular cardiac muscle cell signaling / regulation of atrial cardiac muscle cell action potential / left ventricular cardiac muscle tissue morphogenesis / suramin binding / regulation of AV node cell action potential / regulation of SA node cell action potential / Stimuli-sensing channels / regulation of ventricular cardiac muscle cell action potential / ventricular cardiac muscle cell action potential / positive regulation of sequestering of calcium ion / negative regulation of calcium-mediated signaling / embryonic heart tube morphogenesis / cardiac muscle hypertrophy / Ion homeostasis / negative regulation of insulin secretion involved in cellular response to glucose stimulus / neuronal action potential propagation / negative regulation of release of sequestered calcium ion into cytosol / calcium ion transport into cytosol / insulin secretion involved in cellular response to glucose stimulus / ryanodine-sensitive calcium-release channel activity / response to caffeine / release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / response to redox state / negative regulation of heart rate / cellular response to caffeine / 'de novo' protein folding / calcium ion transmembrane import into cytosol / FK506 binding / response to muscle activity / protein kinase A catalytic subunit binding / protein kinase A regulatory subunit binding / positive regulation of the force of heart contraction / intracellularly gated calcium channel activity / smooth endoplasmic reticulum / smooth muscle contraction / detection of calcium ion / T cell proliferation / positive regulation of heart rate / regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion / calcium channel inhibitor activity / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / Ion homeostasis / response to muscle stretch / release of sequestered calcium ion into cytosol / cellular response to epinephrine stimulus / calcium channel complex / sarcoplasmic reticulum membrane / regulation of heart rate / sarcoplasmic reticulum / protein maturation / sarcomere / calcium channel regulator activity / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / establishment of localization in cell / calcium-mediated signaling / calcium ion transmembrane transport / Stimuli-sensing channels / calcium channel activity / Z disc / intracellular calcium ion homeostasis / calcium ion transport / positive regulation of cytosolic calcium ion concentration / protein refolding / transmembrane transporter binding / response to hypoxia / calmodulin binding / signaling receptor binding / calcium ion binding / protein kinase binding / enzyme binding / protein-containing complex / identical protein binding / membrane / cytoplasm Similarity search - Function | |||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.51 Å | |||||||||||||||||||||
 Authors Authors | Kobayashi T / Tsutsumi A / Kurebayashi N / Kodama M / Kikkawa M / Murayama T / Ogawa H | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular basis for gating of cardiac ryanodine receptor explains the mechanisms for gain- and loss-of function mutations. Authors: Takuya Kobayashi / Akihisa Tsutsumi / Nagomi Kurebayashi / Kei Saito / Masami Kodama / Takashi Sakurai / Masahide Kikkawa / Takashi Murayama / Haruo Ogawa /  Abstract: Cardiac ryanodine receptor (RyR2) is a large Ca release channel in the sarcoplasmic reticulum and indispensable for excitation-contraction coupling in the heart. RyR2 is activated by Ca and RyR2 ...Cardiac ryanodine receptor (RyR2) is a large Ca release channel in the sarcoplasmic reticulum and indispensable for excitation-contraction coupling in the heart. RyR2 is activated by Ca and RyR2 mutations are implicated in severe arrhythmogenic diseases. Yet, the structural basis underlying channel opening and how mutations affect the channel remains unknown. Here, we address the gating mechanism of RyR2 by combining high-resolution structures determined by cryo-electron microscopy with quantitative functional analysis of channels carrying various mutations in specific residues. We demonstrated two fundamental mechanisms for channel gating: interactions close to the channel pore stabilize the channel to prevent hyperactivity and a series of interactions in the surrounding regions is necessary for channel opening upon Ca binding. Mutations at the residues involved in the former and the latter mechanisms cause gain-of-function and loss-of-function, respectively. Our results reveal gating mechanisms of the RyR2 channel and alterations by pathogenic mutations at the atomic level. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33939.map.gz emd_33939.map.gz | 138.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33939-v30.xml emd-33939-v30.xml emd-33939.xml emd-33939.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33939.png emd_33939.png | 124.4 KB | ||
| Others |  emd_33939_half_map_1.map.gz emd_33939_half_map_1.map.gz emd_33939_half_map_2.map.gz emd_33939_half_map_2.map.gz | 113.3 MB 113.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33939 http://ftp.pdbj.org/pub/emdb/structures/EMD-33939 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33939 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33939 | HTTPS FTP |
-Validation report
| Summary document |  emd_33939_validation.pdf.gz emd_33939_validation.pdf.gz | 917.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33939_full_validation.pdf.gz emd_33939_full_validation.pdf.gz | 917.5 KB | Display | |
| Data in XML |  emd_33939_validation.xml.gz emd_33939_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_33939_validation.cif.gz emd_33939_validation.cif.gz | 17 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33939 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33939 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33939 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33939 | HTTPS FTP |
-Related structure data
| Related structure data |  7vmpMC  7vmlC  7vmmC  7vmnC  7vmoC  7vmrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33939.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33939.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of recombinant RyR2 (Ca2 dataset, class 2, open state) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.239 Å | ||||||||||||||||||||||||||||||||||||
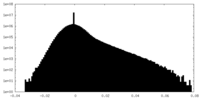
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Structure of recombinant RyR2 (Ca2 dataset, class 2, open state)
| File | emd_33939_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of recombinant RyR2 (Ca2 dataset, class 2, open state) | ||||||||||||
| Projections & Slices |
| ||||||||||||
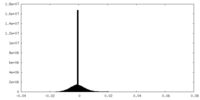
| Density Histograms |
-Half map: Structure of recombinant RyR2 (Ca2 dataset, class 2, open state)
| File | emd_33939_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of recombinant RyR2 (Ca2 dataset, class 2, open state) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Recombinant RyR2 in the presence of Ca2+
| Entire | Name: Recombinant RyR2 in the presence of Ca2+ |
|---|---|
| Components |
|
-Supramolecule #1: Recombinant RyR2 in the presence of Ca2+
| Supramolecule | Name: Recombinant RyR2 in the presence of Ca2+ / type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: in complex with FKBP12.6 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293 Homo sapiens (human) / Recombinant cell: HEK293 |
-Supramolecule #2: Ryanodine receptor 2
| Supramolecule | Name: Ryanodine receptor 2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Supramolecule #3: FKBP1B
| Supramolecule | Name: FKBP1B / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Sugar embedding | Material: buffer |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.51 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 42775 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)