+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SAM-bound MTA1-MTA9-p1-p2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | N6-adenine methylation / MTAc holoenzyme / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmRNA m6A methyltransferase / mRNA m(6)A methyltransferase activity / RNA N6-methyladenosine methyltransferase complex / methylation / nucleus / membrane Similarity search - Function | |||||||||
| Biological species |  Tetrahymena thermophila SB210 (eukaryote) Tetrahymena thermophila SB210 (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Yan JJ / Guan ZY / Liu FQ / Yan XH / Hou MJ / Yin P | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Structural insights into DNA N-adenine methylation by the MTA1 complex. Authors: Junjun Yan / Feiqing Liu / Zeyuan Guan / Xuhui Yan / Xiaohuan Jin / Qiang Wang / Zican Wang / Junjie Yan / Delin Zhang / Zhu Liu / Shan Wu / Ping Yin /  Abstract: N-methyldeoxyadenine (6mA) has recently been reported as a prevalent DNA modification in eukaryotes. The Tetrahymena thermophila MTA1 complex consisting of four subunits, namely MTA1, MTA9, p1, and ...N-methyldeoxyadenine (6mA) has recently been reported as a prevalent DNA modification in eukaryotes. The Tetrahymena thermophila MTA1 complex consisting of four subunits, namely MTA1, MTA9, p1, and p2, is the first identified eukaryotic 6mA methyltransferase (MTase) complex. Unlike the prokaryotic 6mA MTases which have been biochemically and structurally characterized, the operation mode of the MTA1 complex remains largely elusive. Here, we report the cryogenic electron microscopy structures of the quaternary MTA1 complex in S-adenosyl methionine (SAM)-bound (2.6 Å) and S-adenosyl homocysteine (SAH)-bound (2.8 Å) states. Using an AI-empowered integrative approach based on AlphaFold prediction and chemical cross-linking mass spectrometry, we further modeled a near-complete structure of the quaternary complex. Coupled with biochemical characterization, we revealed that MTA1 serves as the catalytic core, MTA1, MTA9, and p1 likely accommodate the substrate DNA, and p2 may facilitate the stabilization of MTA1. These results together offer insights into the molecular mechanism underpinning methylation by the MTA1 complex and the potential diversification of MTases for N-adenine methylation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33854.map.gz emd_33854.map.gz | 49.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33854-v30.xml emd-33854-v30.xml emd-33854.xml emd-33854.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33854.png emd_33854.png | 69.6 KB | ||
| Filedesc metadata |  emd-33854.cif.gz emd-33854.cif.gz | 6.9 KB | ||
| Others |  emd_33854_half_map_1.map.gz emd_33854_half_map_1.map.gz emd_33854_half_map_2.map.gz emd_33854_half_map_2.map.gz | 49 MB 49 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33854 http://ftp.pdbj.org/pub/emdb/structures/EMD-33854 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33854 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33854 | HTTPS FTP |
-Validation report
| Summary document |  emd_33854_validation.pdf.gz emd_33854_validation.pdf.gz | 804.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33854_full_validation.pdf.gz emd_33854_full_validation.pdf.gz | 803.8 KB | Display | |
| Data in XML |  emd_33854_validation.xml.gz emd_33854_validation.xml.gz | 11.9 KB | Display | |
| Data in CIF |  emd_33854_validation.cif.gz emd_33854_validation.cif.gz | 13.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33854 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33854 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33854 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33854 | HTTPS FTP |
-Related structure data
| Related structure data |  7yi9MC  7yi8C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33854.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33854.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33854_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
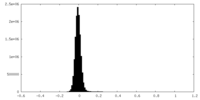
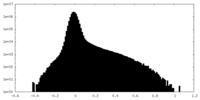
| Density Histograms |
-Half map: #2
| File | emd_33854_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
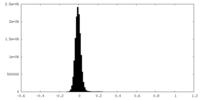
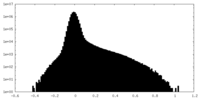
| Density Histograms |
- Sample components
Sample components
-Entire : MTAc holoenzyme
| Entire | Name: MTAc holoenzyme |
|---|---|
| Components |
|
-Supramolecule #1: MTAc holoenzyme
| Supramolecule | Name: MTAc holoenzyme / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) Tetrahymena thermophila SB210 (eukaryote) |
-Macromolecule #1: MTA9
| Macromolecule | Name: MTA9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 |
| Molecular weight | Theoretical: 52.026379 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPKKQEQEP IRLSTRTASK KVDYLQLSNG KLEDFFDDLE EDNKPARNRS RSKKRGRKPL KKADSRSKTP SRVSNARGRS KSLGPRKTY PRKKNLSPDN QLSLLLKWRN DKIPLKSASE TDNKCKVVNV KNIFKSDLSK YGANLQALFI NALWKVKSRK E KEGLNIND ...String: MAPKKQEQEP IRLSTRTASK KVDYLQLSNG KLEDFFDDLE EDNKPARNRS RSKKRGRKPL KKADSRSKTP SRVSNARGRS KSLGPRKTY PRKKNLSPDN QLSLLLKWRN DKIPLKSASE TDNKCKVVNV KNIFKSDLSK YGANLQALFI NALWKVKSRK E KEGLNIND LSNLKIPLSL MKNGILFIWS EKEILGQIVE IMEQKGFTYI ENFSIMFLGL NKCLQSINHK DEDSQNSTAS TN NTNNEAI TSDLTLKDTS KFSDQIQDNH SEDSDQARKQ QTPDDITQKK NKLLKKSSVP SIQKLFEEDP VQTPSVNKPI EKS IEQVTQ EKKFVMNNLD ILKSTDINNL FLRNNYPYFK KTRHTLLMFR RIGDKNQKLE LRHQRTSDVV FEVTDEQDPS KVDT MMKEY VYQMIETLLP KAQFIPGVDK HLKMMELFAS TDNYRPGWIS VIEK UniProtKB: Uncharacterized protein |
-Macromolecule #2: MT-a70 family protein
| Macromolecule | Name: MT-a70 family protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 |
| Molecular weight | Theoretical: 42.696059 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKAVNKKGL RPRKSDSILD HIKNKLDQEF LEDNENGEQS DEDYDQKSLN KAKKPYKKRQ TQNGSELVIS QQKTKAKASA NNKKSAKNS QKLDEEEKIV EEEDLSPQKN GAVSEDDQQQ EASTQEDDYL DRLPKSKKGL QGLLQDIEKR ILHYKQLFFK E QNEIANGK ...String: MSKAVNKKGL RPRKSDSILD HIKNKLDQEF LEDNENGEQS DEDYDQKSLN KAKKPYKKRQ TQNGSELVIS QQKTKAKASA NNKKSAKNS QKLDEEEKIV EEEDLSPQKN GAVSEDDQQQ EASTQEDDYL DRLPKSKKGL QGLLQDIEKR ILHYKQLFFK E QNEIANGK RSMVPDNSIP ICSDVTKLNF QALIDAQMRH AGKMFDVIMM DPPWQLSSSQ PSRGVAIAYD SLSDEKIQNM PI QSLQQDG FIFVWAINAK YRVTIKMIEN WGYKLVDEIT WVKKTVNGKI AKGHGFYLQH AKESCLIGVK GDVDNGRFKK NIA SDVIFS ERRGQSQKPE EIYQYINQLC PNGNYLEIFA RRNNLHDNWV SIGNEL UniProtKB: mRNA m(6)A methyltransferase |
-Macromolecule #3: P1
| Macromolecule | Name: P1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 |
| Molecular weight | Theoretical: 41.602758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLKKGKFQH NQSKSLWNYT LSPGWREEEV KILKSALQLF GIGKWKKIME SGCLPGKSIG QIYMQTQRLL GQQSLGDFMG LQIDLEAVF NQNMKKQDVL RKNNCIINTG DNPTKEERKR RIEQNRKIYG LSAKQIAEIK LPKVKKHAPQ YMTLEDIENE K FTNLEILT ...String: MSLKKGKFQH NQSKSLWNYT LSPGWREEEV KILKSALQLF GIGKWKKIME SGCLPGKSIG QIYMQTQRLL GQQSLGDFMG LQIDLEAVF NQNMKKQDVL RKNNCIINTG DNPTKEERKR RIEQNRKIYG LSAKQIAEIK LPKVKKHAPQ YMTLEDIENE K FTNLEILT HLYNLKAEIV RRLAEQGETI AQPSIIKSLN NLNHNLEQNQ NSNSSTETKV TLEQSGKKKY KVLAIEETEL QN GPIATNS QKKSINGKRK NNRKINSDSE GNEEDISLED IDSQESEINS EEIVEDDEED EQIEEPSKIK KRKKNPEQES EED DIEEDQ EEDELVVNEE EIFEDDDDDE DNQDSSEDDD DDED UniProtKB: Myb-like domain-containing protein |
-Macromolecule #4: Transmembrane protein, putative
| Macromolecule | Name: Transmembrane protein, putative / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 Tetrahymena thermophila SB210 (eukaryote) / Strain: SB210 |
| Molecular weight | Theoretical: 19.886547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKHHHHHHHG AAGTSLYKKA GENLYFQGSM KKNGKSQNQP LDFTQYAKNM RKDLSNQDIC LEDGALNHSY FLTKKGQYWT PLNQKALQR GIELFGVGNW KEINYDEFSG KANIVELELR TCMILGINDI TEYYGKKISE EEQEEIKKSN IAKGKKENKL K DNIYQKLQ QMQ UniProtKB: Transmembrane protein, putative |
-Macromolecule #5: S-ADENOSYLMETHIONINE
| Macromolecule | Name: S-ADENOSYLMETHIONINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: SAM |
|---|---|
| Molecular weight | Theoretical: 398.437 Da |
| Chemical component information |  ChemComp-SAM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)