[English] 日本語
 Yorodumi
Yorodumi- EMDB-33816: Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Tetrahymena ribozyme / second step of self-splicing / conformation 1 / RNA | |||||||||
| Biological species |  Tetrahymena (eukaryote) Tetrahymena (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Li S / Michael ZP / Zhang X / Greg P / Zhang K / Liu L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM. Authors: Shanshan Li / Michael Z Palo / Xiaojing Zhang / Grigore Pintilie / Kaiming Zhang /   Abstract: Group I introns are catalytic RNAs that coordinate two consecutive transesterification reactions for self-splicing. To understand how the group I intron promotes catalysis and coordinates self- ...Group I introns are catalytic RNAs that coordinate two consecutive transesterification reactions for self-splicing. To understand how the group I intron promotes catalysis and coordinates self-splicing reactions, we determine the structures of L-16 Tetrahymena ribozyme in complex with a 5'-splice site analog product and a 3'-splice site analog substrate using cryo-EM. We solve six conformations from a single specimen, corresponding to different splicing intermediates after the first ester-transfer reaction. The structures reveal dynamics during self-splicing, including large conformational changes of the internal guide sequence and the J5/4 junction as well as subtle rearrangements of active-site metals and the hydrogen bond formed between the 2'-OH group of A261 and the N2 group of guanosine substrate. These results help complete a detailed structural and mechanistic view of this paradigmatic group I intron undergoing the second step of self-splicing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33816.map.gz emd_33816.map.gz | 32.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33816-v30.xml emd-33816-v30.xml emd-33816.xml emd-33816.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33816.png emd_33816.png | 50 KB | ||
| Filedesc metadata |  emd-33816.cif.gz emd-33816.cif.gz | 5.1 KB | ||
| Others |  emd_33816_additional_1.map.gz emd_33816_additional_1.map.gz emd_33816_half_map_1.map.gz emd_33816_half_map_1.map.gz emd_33816_half_map_2.map.gz emd_33816_half_map_2.map.gz | 59.4 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33816 http://ftp.pdbj.org/pub/emdb/structures/EMD-33816 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33816 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33816 | HTTPS FTP |
-Related structure data
| Related structure data |  7ygdMC  7yg8C  7yg9C  7ygaC  7ygbC  7ygcC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33816.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33816.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
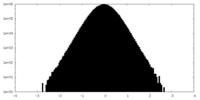
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_33816_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
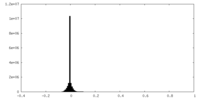
| Density Histograms |
-Half map: #1
| File | emd_33816_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
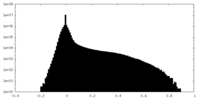
| Density Histograms |
-Half map: #2
| File | emd_33816_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
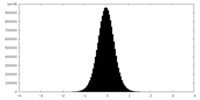
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoi...
| Entire | Name: Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoi...
| Supramolecule | Name: Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Tetrahymena (eukaryote) Tetrahymena (eukaryote) |
| Molecular weight | Theoretical: 130 KDa |
-Macromolecule #1: RNA (5'-R(*UP*CP*G)-3')
| Macromolecule | Name: RNA (5'-R(*UP*CP*G)-3') / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Tetrahymena (eukaryote) Tetrahymena (eukaryote) |
| Molecular weight | Theoretical: 911.596 Da |
| Sequence | String: UCG |
-Macromolecule #2: RNA (5'-R(*CP*C)-3')
| Macromolecule | Name: RNA (5'-R(*CP*C)-3') / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Tetrahymena (eukaryote) Tetrahymena (eukaryote) |
| Molecular weight | Theoretical: 1.788101 KDa |
| Sequence | String: CCCUCU |
-Macromolecule #3: RNA (384-MER)
| Macromolecule | Name: RNA (384-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Tetrahymena (eukaryote) Tetrahymena (eukaryote) |
| Molecular weight | Theoretical: 127.011883 KDa |
| Sequence | String: GGUUUGGAGG GAAAAGUUAU CAGGCAUGCA CCUGGUAGCU AGUCUUUAAA CCAAUAGAUU GCAUCGGUUU AAAAGGCAAG ACCGUCAAA UUGCGGGAAA GGGGUCAACA GCCGUUCAGU ACCAAGUCUC AGGGGAAACU UUGAGAUGGC CUUGCAAAGG G UAUGGUAA ...String: GGUUUGGAGG GAAAAGUUAU CAGGCAUGCA CCUGGUAGCU AGUCUUUAAA CCAAUAGAUU GCAUCGGUUU AAAAGGCAAG ACCGUCAAA UUGCGGGAAA GGGGUCAACA GCCGUUCAGU ACCAAGUCUC AGGGGAAACU UUGAGAUGGC CUUGCAAAGG G UAUGGUAA UAAGCUGACG GACAUGGUCC UAACCACGCA GCCAAGUCCU AAGUCAACAG AUCUUCUGUU GAUAUGGAUG CA GUUCACA GACUAAAUGU CGGUCGGGGA AGAUGUAUUC UUCUCAUAAG AUAUAGUCGG ACCUCUCCUU AAUGGGAGCU AGC GGAUGA AGUGAUGCAA CACUGGAGCC GCUGGGAACU AAUUUGUAUG CGAAAGUAUA UUGAUUAGUU UUGGAGU GENBANK: GENBANK: X54512.1 |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.6 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 52.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)