+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of EBOV L-VP35-RNA complex (state2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polymerase / complex / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IKBKE activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF7 activity / GDP polyribonucleotidyltransferase / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / Transferases; Transferring one-carbon groups; Methyltransferases / virion component / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IKBKE activity / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of IRF7 activity / GDP polyribonucleotidyltransferase / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / Transferases; Transferring one-carbon groups; Methyltransferases / virion component / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / symbiont-mediated suppression of host toll-like receptor signaling pathway / host cell cytoplasm / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA-directed RNA polymerase / RNA-directed RNA polymerase activity / GTPase activity / RNA binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
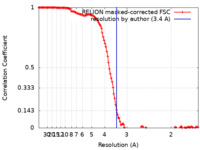
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Shi Y / Yuan B / Peng Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structure of the Ebola virus polymerase complex. Authors: Bin Yuan / Qi Peng / Jinlong Cheng / Min Wang / Jin Zhong / Jianxun Qi / George F Gao / Yi Shi /  Abstract: Filoviruses, including Ebola virus, pose an increasing threat to the public health. Although two therapeutic monoclonal antibodies have been approved to treat the Ebola virus disease, there are no ...Filoviruses, including Ebola virus, pose an increasing threat to the public health. Although two therapeutic monoclonal antibodies have been approved to treat the Ebola virus disease, there are no approved broadly reactive drugs to control diverse filovirus infection. Filovirus has a large polymerase (L) protein and the cofactor viral protein 35 (VP35), which constitute the basic functional unit responsible for virus genome RNA synthesis. Owing to its conservation, the L-VP35 polymerase complex is a promising target for broadly reactive antiviral drugs. Here we determined the structure of Ebola virus L protein in complex with tetrameric VP35 using cryo-electron microscopy (state 1). Structural analysis revealed that Ebola virus L possesses a filovirus-specific insertion element that is essential for RNA synthesis, and that VP35 interacts extensively with the N-terminal region of L by three protomers of the VP35 tetramer. Notably, we captured the complex structure in a second conformation with the unambiguous priming loop and supporting helix away from polymerase active site (state 2). Moreover, we demonstrated that the century-old drug suramin could inhibit the activity of the Ebola virus polymerase in an enzymatic assay. The structure of the L-VP35-suramin complex reveals that suramin can bind at the highly conserved NTP entry channel to prevent substrates from entering the active site. These findings reveal the mechanism of Ebola virus replication and may guide the development of more powerful anti-filovirus drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33776.map.gz emd_33776.map.gz | 12.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33776-v30.xml emd-33776-v30.xml emd-33776.xml emd-33776.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33776_fsc.xml emd_33776_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_33776.png emd_33776.png | 42.5 KB | ||
| Filedesc metadata |  emd-33776.cif.gz emd-33776.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33776 http://ftp.pdbj.org/pub/emdb/structures/EMD-33776 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33776 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33776 | HTTPS FTP |
-Related structure data
| Related structure data |  7yesMC  7yerC  7yetC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33776.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33776.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : The complex of EBOV L-VP35
| Entire | Name: The complex of EBOV L-VP35 |
|---|---|
| Components |
|
-Supramolecule #1: The complex of EBOV L-VP35
| Supramolecule | Name: The complex of EBOV L-VP35 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 252.863734 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MATQHTQYPD ARLSSPIVLD QCDLVTRACG LYSSYSLNPQ LRNCKLPKHI YRLKYDVTVT KFLSDVPVAT LPIDFIVPIL LKALSGNGF CPVEPRCQQF LDEIIKYTMQ DALFLKYYLK NVGAQEDCVD DHFQEKILSS IQGNEFLHQM FFWYDLAILT R RGRLNRGN ...String: MATQHTQYPD ARLSSPIVLD QCDLVTRACG LYSSYSLNPQ LRNCKLPKHI YRLKYDVTVT KFLSDVPVAT LPIDFIVPIL LKALSGNGF CPVEPRCQQF LDEIIKYTMQ DALFLKYYLK NVGAQEDCVD DHFQEKILSS IQGNEFLHQM FFWYDLAILT R RGRLNRGN SRSTWFVHDD LIDILGYGDY VFWKIPISLL PLNTQGIPHA AMDWYQTSVF KEAVQGHTHI VSVSTADVLI MC KDLITCR FNTTLISKIA EVEDPVCSDY PNFKIVSMLY QSGDYLLSIL GSDGYKIIKF LEPLCLAKIQ LCSKYTERKG RFL TQMHLA VNHTLEEITE IRALKPSQAH KIREFHRTLI RLEMTPQQLC ELFSIQKHWG HPVLHSETAI QKVKKHATVL KALR PIVIF ETYCVFKYSI AKHYFDSQGS WYSVTSDRNL TPGLNSYIKR NQFPPLPMIK ELLWEFYHLD HPPLFSTKII SDLSI FIKD RATAVERTCW DAVFEPNVLG YNPPHKFSTK RVPEQFLEQE NFSIENVLSY AQKLEYLLPQ YRNFSFSLKE KELNVG RTF GKLPYPTRNV QTLCEALLAD GLAKAFPSNM MVVTEREQKE SLLHQASWHH TSDDFGEHAT VRGSSFVTDL EKYNLAF RY EFTAPFIEYC NRCYGVKNVF NWMHYTIPQC YMHVSDYYNP PHNLTLENRN NPPEGPSSYR GHMGGIEGLQ QKLWTSIS C AQISLVEIKT GFKLRSAVMG DNQCITVLSV FPLETDADEQ EQSAEDNAAR VAASLAKVTS ACGIFLKPDE TFVHSGFIY FGKKQYLNGV QLPQSLKTAT RMAPLSDAIF DDLQGTLASI GTAFERSISE TRHIFPCRIT AAFHTFFSVR ILQYHHLGFN KGFDLGQLT LGKPLDFGTI SLALAVPQVL GGLSFLNPEK CFYRNLGDPV TSGLFQLKTY LRMIEMDDLF LPLIAKNPGN C TAIDFVLN PSGLNVPGSQ DLTSFLRQIV RRTITLSAKN KLINTLFHAS ADFEDEMVCK WLLSSTPVMS RFAADIFSRT PS GKRLQIL GYLEGTRTLL ASKIINNNTE TPVLDRLRKI TLQRWSLWFS YLDHCDNILA EALTQITCTV DLAQILREYS WAH ILEGRP LIGATLPCMI EQFKVVWLKP YEQCPQCSNA KQPGGKPFVS VAVKKHIVSA WPNASRISWT IGDGIPYIGS RTED KIGQP AIKPKCPSAA LREAIELASR LTWVTQGSSN SDLLIKPFLE ARVNLSVQEI LQMTPSHYSG NIVHRYNDQY SPHSF MANR MSNSATRLIV STNTLGEFSG GGQSARDSNI IFQNVINYAV ALFDIKFRNT EATDIQYNRA HLHLTKCCTR EVPAQY LTY TSTLDLDLTR YRENELIYDN NPLKGGLNCN ISFDNPFFQG KQLNIIEDDL IRLPHLSGWE LAKTIMQSII SDSNNSS TD PISSGETRSF TTHFLTYPKI GLLYSFGAFV SYYLGNTILR TKKLTLDNFL YYLTTQIHNL PHRSLRILKP TFKHASVM S RLMSIDPHFS IYIGGAAGDR GLSDAARLFL RTSISSFLTF VKEWIINRGT IVPLWIVYPL EGQNPTPVNN FLHQIVELL VHDSSRHQAF KTTINDHVHP HDNLVYTCKS TASNFFHASL AYWRSRHRNS NRKDLTRNSS TGSSTNNSDG HIKRSQEQTT RDPHDGTER SLVLQMSHEI KRTTIPQENT HQGPSFQSFL SDSACGTANP KLNFDRSRHN VKSQDHNSAS KREGHQIISH R LVLPFFTL SQGTRQLTSS NESQTQDEIS KYLRQLRSVI DTTVYCRFTG IVSSMHYKLD EVLWEIENFK SAVTLAEGEG AG ALLLIQK YQVKTLFFNT LATESSIESE IVSGMTTPRM LLPVMSKFHN DQIEIILNNS ASQITDITNP TWFKDQRARL PRQ VEVITM DAETTENINR SKLYEAVHKL ILHHVDPSVL KAVVLKVFLS DTEGMLWLND NLAPFFATGY LIKPITSSAR SSEW YLCLT NFLSTTRKMP HQNHLSCKQV ILTALQLQIQ RSPYWLSHLT QYADCDLHLS YIRLGFPSLE KVLYHRYNLV DSKRG PLVS VTQHLAHLRA EIRELTNDYN QQRQSRTQTY HFIRTAKGRI TKLVNDYLKF FLIVQALKHN GTWQAEFKKL PELISV CNR FYHIRDCNCE ERFLVQTLYL HRMQDSEVKL IERLTGLLSL FPDGLYRFD UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: VP35 of EBOV L-VP35 complex
| Macromolecule | Name: VP35 of EBOV L-VP35 complex / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.48943 KDa |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MTTRTKGRGH TVATTQNDRM PGPELSGWIS EQLMTGRIPV NDIFCDIENN PGLCYASQMQ QTKPNPKMRN SQTQTDPICN HSFEEVVQT LASLATVVQQ QTIASESLEQ RITSLENGLK PVYDMAKTIS SLNRVCAEMV AKYDLLVMTT GRATATAAAT E AYWAEHGQ ...String: MTTRTKGRGH TVATTQNDRM PGPELSGWIS EQLMTGRIPV NDIFCDIENN PGLCYASQMQ QTKPNPKMRN SQTQTDPICN HSFEEVVQT LASLATVVQQ QTIASESLEQ RITSLENGLK PVYDMAKTIS SLNRVCAEMV AKYDLLVMTT GRATATAAAT E AYWAEHGQ PPPGPSLYEE SAIRGKIESR DETVPQSVRE AFNNLDSTTS LTEENFGKPD ISAKDLRNIM YDHLPGFGTA FH QLVQVIC KLGKDSNSLD IIHAEFQASL AEGDSPQCAL IQITKRVPIF QDAAPPVIHI RSRGDIPRAC QKSLRPVPPS PKI DRGWVC VFQLQDGKTL GLKI UniProtKB: Polymerase cofactor VP35 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)