+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 2-APB bound state of mTRPV2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mTRPV2 / 2-APB / STRUCTURAL PROTEIN / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgrowth cone membrane / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / axonal growth cone / positive regulation of axon extension / monoatomic cation channel activity / calcium channel activity / melanosome / positive regulation of cold-induced thermogenesis ...growth cone membrane / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / axonal growth cone / positive regulation of axon extension / monoatomic cation channel activity / calcium channel activity / melanosome / positive regulation of cold-induced thermogenesis / cell body / axon / cell surface / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Su N | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands. Authors: Nannan Su / Wenxuan Zhen / Heng Zhang / Lingyi Xu / Yitian Jin / Xiaoying Chen / Cheng Zhao / Qinrui Wang / Xinyan Wang / Shaowei Li / Han Wen / Wei Yang / Jiangtao Guo / Fan Yang /  Abstract: The transient receptor potential vanilloid 2 (TRPV2) ion channel is a polymodal receptor widely involved in many physiological and pathological processes. Despite many TRPV2 modulators being ...The transient receptor potential vanilloid 2 (TRPV2) ion channel is a polymodal receptor widely involved in many physiological and pathological processes. Despite many TRPV2 modulators being identified, whether and how TRPV2 is regulated by endogenous lipids remains elusive. Here, we report an endogenous cholesterol molecule inside the vanilloid binding pocket (VBP) of TRPV2, with a 'head down, tail up' configuration, resolved at 3.2 Å using cryo-EM. Cholesterol binding antagonizes ligand activation of TRPV2, which is removed from VBP by methyl-β-cyclodextrin (MβCD) as resolved at 2.9 Å. We also observed that estradiol (E2) potentiated TRPV2 activation by 2-aminoethoxydiphenyl borate (2-APB), a classic tool compound for TRP channels. Our cryo-EM structures (resolved at 2.8-3.3 Å) further suggest how E2 disturbed cholesterol binding and how 2-APB bound within the VBP with E2 or without both E2 and endogenous cholesterol, respectively. Therefore, our study has established the structural basis for ligand recognition of the inhibitory endogenous cholesterol and excitatory exogenous 2-APB in TRPV2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33774.map.gz emd_33774.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33774-v30.xml emd-33774-v30.xml emd-33774.xml emd-33774.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33774.png emd_33774.png | 70.7 KB | ||
| Masks |  emd_33774_msk_1.map emd_33774_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33774.cif.gz emd-33774.cif.gz | 6.4 KB | ||
| Others |  emd_33774_half_map_1.map.gz emd_33774_half_map_1.map.gz emd_33774_half_map_2.map.gz emd_33774_half_map_2.map.gz | 14 MB 14 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33774 http://ftp.pdbj.org/pub/emdb/structures/EMD-33774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33774 | HTTPS FTP |
-Validation report
| Summary document |  emd_33774_validation.pdf.gz emd_33774_validation.pdf.gz | 831 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33774_full_validation.pdf.gz emd_33774_full_validation.pdf.gz | 830.6 KB | Display | |
| Data in XML |  emd_33774_validation.xml.gz emd_33774_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_33774_validation.cif.gz emd_33774_validation.cif.gz | 13.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33774 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33774 | HTTPS FTP |
-Related structure data
| Related structure data |  7yepMC  7xemC  7xeoC  7xerC  7xeuC  7xevC  7xewC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33774.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33774.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
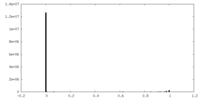
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33774_msk_1.map emd_33774_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
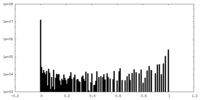
| Density Histograms |
-Half map: #2
| File | emd_33774_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
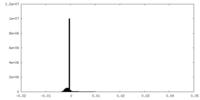
| Density Histograms |
-Half map: #1
| File | emd_33774_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
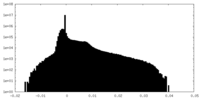
| Density Histograms |
- Sample components
Sample components
-Entire : complex of mTRPV2 and 2-APB
| Entire | Name: complex of mTRPV2 and 2-APB |
|---|---|
| Components |
|
-Supramolecule #1: complex of mTRPV2 and 2-APB
| Supramolecule | Name: complex of mTRPV2 and 2-APB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Transient receptor potential cation channel subfamily V member 2
| Macromolecule | Name: Transient receptor potential cation channel subfamily V member 2 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 86.058195 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTSASNPPAF RLETSDGDEE GSAEVNKGKN EPPPMESPFQ GEDRNFSPQI KVNLNYRKGL GPSQQDPNRF DRDRLFSVVS RGVPEELTG LLEYLRRTSK YLTDSAYTEG STGKTCLMKA VLNLQDGVNA CILPLLQIDR DSGNPQPLVN AQCTDEFYRG H SALHIAIE ...String: MTSASNPPAF RLETSDGDEE GSAEVNKGKN EPPPMESPFQ GEDRNFSPQI KVNLNYRKGL GPSQQDPNRF DRDRLFSVVS RGVPEELTG LLEYLRRTSK YLTDSAYTEG STGKTCLMKA VLNLQDGVNA CILPLLQIDR DSGNPQPLVN AQCTDEFYRG H SALHIAIE KRSLWCVKLL VENGANVHIR ACGRFFQKHQ GTCFYFGELP LSLAACTKQW DVVTYLLENP HQPASLEATD SL GNTVLHA LVMIADNSPE NSALVIHMYD SLLQMGARLC PTVQLEDICN HQGLTPLKLA AKEGKIEIFR HILQREFSGL YQP LSRKFT EWCYGPVRVS LYDLSSVDSW EKNSVLEIIA FHCKSPHRHR MVVLEPLNKL LQEKWDRLIP RFFFNFACYL VYMI IFTIV AYHQPSLEQP AIPSSKATFG DSMLLLGHIL ILLGGIYLLL GQLWYFWRRR LFIWISFMDS YFEILFLVQA LLTVL SQVL RFVETEWYLP LLVSSLVLGW LNLLYYTRGF QHTGIYSVMI QKVILRDLLR FLLVYLVFLF GFAVALVSLS REARSP KAP EDSNTTVTEK PTLGQEEEPV PYGGILDASL ELFKFTIGMG ELAFQEQLRF RGVVLLLLLA YVLLTYVLLL NMLIALM SE TVNSVATDSW SIWKLQKAIS VLEMENGYWW CRRKRHRAGR LLKVGTKGDG IPDERWCFRV EEVNWAAWEK TLPTLSED P SGAGITGYKK NPTSKPGKNS ASEEDHLPLQ VLQSH UniProtKB: Transient receptor potential cation channel subfamily V member 2 |
-Macromolecule #2: 2-aminoethyl diphenylborinate
| Macromolecule | Name: 2-aminoethyl diphenylborinate / type: ligand / ID: 2 / Number of copies: 4 / Formula: FZ4 |
|---|---|
| Molecular weight | Theoretical: 225.094 Da |
| Chemical component information | 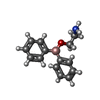 ChemComp-FZ4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 6.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)