+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a eukaryotic ZnT8 in the presence of zinc | |||||||||
 Map data Map data | xtZnT8-Zn2 in the presence of 1 mM Zn2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Zinc transporter / SLC30 / ZnT / CDF / Proton-coupled antiporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationInsulin processing / Zinc efflux and compartmentalization by the SLC30 family / zinc:proton antiporter activity / zinc ion import into organelle / transport vesicle membrane / secretory granule membrane / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species | ||||||||||
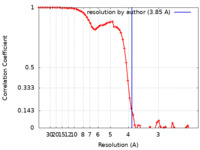
| Method | single particle reconstruction / cryo EM / Resolution: 3.85 Å | |||||||||
 Authors Authors | Zhang S / Fu C / Luo Y / Sun Z / Su Z / Zhou X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2023 Journal: J Struct Biol / Year: 2023Title: Cryo-EM structure of a eukaryotic zinc transporter at a low pH suggests its Zn-releasing mechanism. Authors: Senfeng Zhang / Chunting Fu / Yongbo Luo / Qingrong Xie / Tong Xu / Ziyi Sun / Zhaoming Su / Xiaoming Zhou /  Abstract: Zinc transporter 8 (ZnT8) is mainly expressed in pancreatic islet β cells and is responsible for H-coupled uptake (antiport) of Zn into the lumen of insulin secretory granules. Structures of human ...Zinc transporter 8 (ZnT8) is mainly expressed in pancreatic islet β cells and is responsible for H-coupled uptake (antiport) of Zn into the lumen of insulin secretory granules. Structures of human ZnT8 and its prokaryotic homolog YiiP have provided structural basis for constructing a plausible transport cycle for Zn. However, the mechanistic role that protons play in the transport process remains unclear. Here we present a lumen-facing cryo-EM structure of ZnT8 from Xenopus tropicalis (xtZnT8) in the presence of Zn at a luminal pH (5.5). Compared to a Zn-bound xtZnT8 structure at a cytosolic pH (7.5), the low-pH structure displays an empty transmembrane Zn-binding site with a disrupted coordination geometry. Combined with a Zn-binding assay our data suggest that protons may disrupt Zn coordination at the transmembrane Zn-binding site in the lumen-facing state, thus facilitating Zn release from ZnT8 into the lumen. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33619.map.gz emd_33619.map.gz | 24.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33619-v30.xml emd-33619-v30.xml emd-33619.xml emd-33619.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33619_fsc.xml emd_33619_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_33619.png emd_33619.png | 111.8 KB | ||
| Filedesc metadata |  emd-33619.cif.gz emd-33619.cif.gz | 5.9 KB | ||
| Others |  emd_33619_half_map_1.map.gz emd_33619_half_map_1.map.gz emd_33619_half_map_2.map.gz emd_33619_half_map_2.map.gz | 23.7 MB 23.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33619 http://ftp.pdbj.org/pub/emdb/structures/EMD-33619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33619 | HTTPS FTP |
-Validation report
| Summary document |  emd_33619_validation.pdf.gz emd_33619_validation.pdf.gz | 706.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33619_full_validation.pdf.gz emd_33619_full_validation.pdf.gz | 706.1 KB | Display | |
| Data in XML |  emd_33619_validation.xml.gz emd_33619_validation.xml.gz | 13.7 KB | Display | |
| Data in CIF |  emd_33619_validation.cif.gz emd_33619_validation.cif.gz | 17.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33619 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33619 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33619 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33619 | HTTPS FTP |
-Related structure data
| Related structure data |  7y5gMC  7y5hC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33619.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33619.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | xtZnT8-Zn2 in the presence of 1 mM Zn2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33619_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33619_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : xtZnT8 dimer complexed with zinc
| Entire | Name: xtZnT8 dimer complexed with zinc |
|---|---|
| Components |
|
-Supramolecule #1: xtZnT8 dimer complexed with zinc
| Supramolecule | Name: xtZnT8 dimer complexed with zinc / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: Zinc transporter 8
| Macromolecule | Name: Zinc transporter 8 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 41.539031 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MKGSEEAYLV SDKATKMYSL TKDSEKNHPS KPPLQDEENP QSKYHCHNNN KKAYDARQRE QTFAKKKLCI ASLICFVFIS AEIVGGYIA GSLAVVTDAA HLLVDLSSFF ISLCSLWLSS KSSTTRLTFG WHRAEILGAL MSVITIWLVT GVLVYLACER L IRPDYTID ...String: MKGSEEAYLV SDKATKMYSL TKDSEKNHPS KPPLQDEENP QSKYHCHNNN KKAYDARQRE QTFAKKKLCI ASLICFVFIS AEIVGGYIA GSLAVVTDAA HLLVDLSSFF ISLCSLWLSS KSSTTRLTFG WHRAEILGAL MSVITIWLVT GVLVYLACER L IRPDYTID GTVMLITSAC ALGANLVLAL ILHQSGHGHS HAGGKHEHMA SEYKPQTNAS IRAAFIHVIG DLFQSISVLI SA LIIYFKP EYKMADPICT FIFSIFVLIT TVTVLRDLLT VLMEGTPRGI HYSDVKQSIL AVDGVKSVHS LHLWALTMNQ VIL SAHIAT DIVGESKRIL KDVTQNVFAR FPFHSVTIQV EPIEDQSPEC MFCYEPTQ UniProtKB: Proton-coupled zinc antiporter SLC30A8 |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM 2-mercaptoethanol, 0.5 mM DDM and 1 mM ZnSO4 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 2-3 s before plunging. |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -1.7 µm / Nominal defocus min: -1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)