[English] 日本語
 Yorodumi
Yorodumi- EMDB-33600: Cryo-EM structure of the SARS-CoV-2 spike glycoprotein in complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein in complex with all-trans retinoic acid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VIRAL PROTEIN-INHIBITOR COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Ye X / Lin W | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: mBio / Year: 2022 Journal: mBio / Year: 2022Title: A Retinol Derivative Inhibits SARS-CoV-2 Infection by Interrupting Spike-Mediated Cellular Entry. Authors: Liangqin Tong / Lin Wang / Shumin Liao / Xiaoping Xiao / Jing Qu / Chunli Wu / Yibin Zhu / Wanbo Tai / Yanhong Huang / Penghua Wang / Liang Li / Renli Zhang / Ye Xiang / Gong Cheng /   Abstract: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of the global pandemic and life-threatening coronavirus disease 2019 (COVID-19). Although vaccines and ...Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiological agent of the global pandemic and life-threatening coronavirus disease 2019 (COVID-19). Although vaccines and therapeutic antibodies are available, their efficacy is continuously undermined by rapidly emerging SARS-CoV-2 variants. Here, we found that all- retinoic acid (ATRA), a vitamin A (retinol) derivative, showed potent antiviral activity against all SARS-CoV-2 variants in both human cell lines and human organoids of the lower respiratory tract. Mechanistically, ATRA directly binds in a deep hydrophobic pocket of the receptor binding domain (RBD) located on the top of the SARS-CoV-2 spike protein (S) trimer. The bound ATRA mediates strong interactions between the "down" RBDs and locks most of the S trimers in an RBD "all-down" and ACE2-inaccessible inhibitory conformation. In summary, our results reveal the pharmacological biotargets and structural mechanism of ATRA and other retinoids in SARS-CoV-2 infection and suggest that ATRA and its derivatives could be potential hit compounds against a broad spectrum of coronaviruses. Retinoids, a group of compounds including vitamin A and its active metabolite all- retinoic acid (ATRA), regulate serial physiological activity in multiple organ systems, such as cell growth, differentiation, and apoptosis. The ATRA analogues reported to date include more than 4,000 natural and synthetic molecules that are structurally and/or functionally related to ATRA. Here, we found that ATRA showed potent antiviral activity against all SARS-CoV-2 variants by directly binding in a deep hydrophobic pocket of the receptor binding domain (RBD) located on top of the SARS-CoV-2 spike protein (S) trimer. The bound ATRA mediates strong interactions between the "down" RBDs and locks most of the S trimers in an RBD "all-down" and ACE2-inaccessible inhibitory conformation, suggesting the pharmacological feasibility of using ATRA or its derivatives as a remedy for and prevention of COVID-19 disease. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33600.map.gz emd_33600.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33600-v30.xml emd-33600-v30.xml emd-33600.xml emd-33600.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33600.png emd_33600.png | 56.1 KB | ||
| Filedesc metadata |  emd-33600.cif.gz emd-33600.cif.gz | 6.1 KB | ||
| Others |  emd_33600_half_map_1.map.gz emd_33600_half_map_1.map.gz emd_33600_half_map_2.map.gz emd_33600_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33600 http://ftp.pdbj.org/pub/emdb/structures/EMD-33600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33600 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33600 | HTTPS FTP |
-Related structure data
| Related structure data |  7y42MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33600.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33600.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33600_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33600_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike glycoprotein in complex with ATRA
| Entire | Name: SARS-CoV-2 spike glycoprotein in complex with ATRA |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike glycoprotein in complex with ATRA
| Supramolecule | Name: SARS-CoV-2 spike glycoprotein in complex with ATRA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 Details: "GSAS" substitution at the furin cleavage site (residues 682-685) Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 124.168508 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AYTNSFTRGV YYPDKVFRSS VLHSTQDLFL PFFSNVTWFH AIHVSGTNGT KRFDNPVLPF NDGVYFASTE KSNIIRGWIF GTTLDSKTQ SLLIVNNATN VVIKVCEFQF CNDPFLGVYY HKNNKSWMES EFRVYSSANN FTFEYVSQPF LMDLEGKQGN F KNLREFVF ...String: AYTNSFTRGV YYPDKVFRSS VLHSTQDLFL PFFSNVTWFH AIHVSGTNGT KRFDNPVLPF NDGVYFASTE KSNIIRGWIF GTTLDSKTQ SLLIVNNATN VVIKVCEFQF CNDPFLGVYY HKNNKSWMES EFRVYSSANN FTFEYVSQPF LMDLEGKQGN F KNLREFVF KNIDGYFKIY SKHTPINLVR DLPQGFSALE PLVDLPIGIN ITRFQTLLAL HRSYLTPGDS SSGWTAGAAA YY VGYLQPR TFLLKYNENG TITDAVDCAL DPLSETKCTL KSFTVEKGIY QTSNFRVQPT ESIVRFPNIT NLCPFGEVFN ATR FASVYA WNRKRISNCV ADYSVLYNSA SFSTFKCYGV SPTKLNDLCF TNVYADSFVI RGDEVRQIAP GQTGKIADYN YKLP DDFTG CVIAWNSNNL DSKVGGNYNY LYRLFRKSNL KPFERDISTE IYQAGSTPCN GVEGFNCYFP LQSYGFQPTN GVGYQ PYRV VVLSFELLHA PATVCGPKKS TNLVKNKCVN FNFNGLTGTG VLTESNKKFL PFQQFGRDIA DTTDAVRDPQ TLEILD ITP CSFGGVSVIT PGTNTSNQVA VLYQDVNCTE VPVAIHADQL TPTWRVYSTG SNVFQTRAGC LIGAEHVNNS YECDIPI GA GICASYQTQT NSPRRARSVA SQSIIAYTMS LGAENSVAYS NNSIAIPTNF TISVTTEILP VSMTKTSVDC TMYICGDS T ECSNLLLQYG SFCTQLNRAL TGIAVEQDKN TQEVFAQVKQ IYKTPPIKDF GGFNFSQILP DPSKPSKRSF IEDLLFNKV TLADAGFIKQ YGDCLGDIAA RDLICAQKFN GLTVLPPLLT DEMIAQYTSA LLAGTITSGW TFGAGAALQI PFAMQMAYRF NGIGVTQNV LYENQKLIAN QFNSAIGKIQ DSLSSTASAL GKLQDVVNQN AQALNTLVKQ LSSNFGAISS VLNDILSRLD P PEAEVQID RLITGRLQSL QTYVTQQLIR AAEIRASANL AATKMSECVL GQSKRVDFCG KGYHLMSFPQ SAPHGVVFLH VT YVPAQEK NFTTAPAICH DGKAHFPREG VFVSNGTHWF VTQRNFYEPQ IITTDNTFVS GNCDVVIGIV NNTVYDPLQP ELD S UniProtKB: Spike glycoprotein |
-Macromolecule #2: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 2 / Number of copies: 27 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #3: RETINOIC ACID
| Macromolecule | Name: RETINOIC ACID / type: ligand / ID: 3 / Number of copies: 3 / Formula: REA |
|---|---|
| Molecular weight | Theoretical: 300.435 Da |
| Chemical component information | 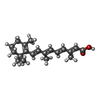 ChemComp-3KV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Sugar embedding | Material: vitrified ice |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: EMDB MAP EMDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.45 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 66436 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































