[English] 日本語
 Yorodumi
Yorodumi- EMDB-33570: 5.0 angstrom cryo-EM structure of transmembrane regions of mouse ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 5.0 angstrom cryo-EM structure of transmembrane regions of mouse Basigin/MCT1 in complex with antibody 6E7F1 | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transporter / glucose metabolism / monocarboxylic acid transport / alpha helical TMs / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationProton-coupled monocarboxylate transport / Aspirin ADME / behavioral response to nutrient / plasma membrane lactate transport / pyruvate transmembrane transport / lactate transmembrane transporter activity / lactate:proton symporter activity / Basigin interactions / solute:proton symporter activity / positive regulation of matrix metallopeptidase secretion ...Proton-coupled monocarboxylate transport / Aspirin ADME / behavioral response to nutrient / plasma membrane lactate transport / pyruvate transmembrane transport / lactate transmembrane transporter activity / lactate:proton symporter activity / Basigin interactions / solute:proton symporter activity / positive regulation of matrix metallopeptidase secretion / succinate transmembrane transporter activity / acrosomal membrane / Degradation of the extracellular matrix / dendrite self-avoidance / Integrin cell surface interactions / endothelial tube morphogenesis / pyruvate catabolic process / cell-cell adhesion mediator activity / response to mercury ion / neural retina development / photoreceptor cell maintenance / centrosome cycle / neuron projection extension / response to food / odontogenesis of dentin-containing tooth / D-mannose binding / homophilic cell-cell adhesion / decidualization / lateral plasma membrane / positive regulation of vascular endothelial growth factor production / photoreceptor outer segment / response to cAMP / neutrophil chemotaxis / photoreceptor inner segment / positive regulation of endothelial cell migration / regulation of insulin secretion / embryo implantation / axon guidance / protein localization to plasma membrane / lipid metabolic process / sarcolemma / response to peptide hormone / positive regulation of interleukin-6 production / signaling receptor activity / glucose homeostasis / virus receptor activity / angiogenesis / spermatogenesis / basolateral plasma membrane / positive regulation of viral entry into host cell / apical plasma membrane / endosome / axon / synapse / centrosome / endoplasmic reticulum membrane / mitochondrion / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.03 Å | |||||||||
 Authors Authors | Zhang H / Yang X / Xue Y / Huang Y / Mo X / Li N / Gao N / Li X / Wang S / Gao Y / Liao J | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin-2 Authors: Zhang H / Yang X / Xue Y / Huang Y / Mo Y / Zhang H / Li N / Gao N / Li X / Wang S / Gao Y / Liao J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33570.map.gz emd_33570.map.gz | 49.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33570-v30.xml emd-33570-v30.xml emd-33570.xml emd-33570.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33570.png emd_33570.png | 79.2 KB | ||
| Masks |  emd_33570_msk_1.map emd_33570_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33570.cif.gz emd-33570.cif.gz | 7 KB | ||
| Others |  emd_33570_half_map_1.map.gz emd_33570_half_map_1.map.gz emd_33570_half_map_2.map.gz emd_33570_half_map_2.map.gz | 48.9 MB 48.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33570 http://ftp.pdbj.org/pub/emdb/structures/EMD-33570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33570 | HTTPS FTP |
-Related structure data
| Related structure data |  7y1qMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33570.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33570.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33570_msk_1.map emd_33570_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_33570_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_33570_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
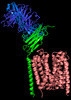
-Entire : Ternary complex of transmembrane regions of Basigin with monocarb...
| Entire | Name: Ternary complex of transmembrane regions of Basigin with monocarboxylate transporter 1 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of transmembrane regions of Basigin with monocarb...
| Supramolecule | Name: Ternary complex of transmembrane regions of Basigin with monocarboxylate transporter 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Ternary complex of transmembrane regions of Basigin with monocarboxylate transporter 1, extracellular region of Basigin is missed due to high flexibility |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: Monocarboxylate transporter 1
| Macromolecule | Name: Monocarboxylate transporter 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.308883 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPPAIGGPVG YTPPDGGWGW AVLVGAFISI GFSYAFPKSI TVFFKEIEVI FSATTSEVSW ISSIMLAVMY AGGPISSILV NKYGSRPVM IAGGCLSGCG LIAASFCNTV QELYLCIGVI GGLGLAFNLN PALTMIGKYF YKKRPLANGL AMAGSPVFLS T LAPLNQAF ...String: MPPAIGGPVG YTPPDGGWGW AVLVGAFISI GFSYAFPKSI TVFFKEIEVI FSATTSEVSW ISSIMLAVMY AGGPISSILV NKYGSRPVM IAGGCLSGCG LIAASFCNTV QELYLCIGVI GGLGLAFNLN PALTMIGKYF YKKRPLANGL AMAGSPVFLS T LAPLNQAF FDIFDWRGSF LILGGLLLNC CVAGSLMRPI GPEQVKLEKL KSKESLQEAG KSDANTDLIG GSPKGEKLSV FQ TINKFLD LSLFTHRGFL LYLSGNVVMF FGLFTPLVFL SSYGKSKDFS SEKSAFLLSI LAFVDMVARP SMGLAANTKW IRP RIQYFF AASVVANGVC HLLAPLSTTY VGFCVYAGVF GFAFGWLSSV LFETLMDLIG PQRFSSAVGL VTIVECCPVL LGPP LLGRL NDMYGDYKYT YWACGVILII AGIYLFIGMG INYRLLAKEQ KAEEKQKREG KEDEASTDVD EKPKETMKAA QSPQQ HSSG DPTEEESPV UniProtKB: Monocarboxylate transporter 1 |
-Macromolecule #2: Isoform 2 of Basigin
| Macromolecule | Name: Isoform 2 of Basigin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.707447 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAALLLALA FTLLSGQGAC AAAGTIQTSV QEVNSKTQLT CSLNSSGVDI VGHRWMRGGK VLQEDTLPDL HTKYIVDADD RSGEYSCIF LPEPVGRSEI NVEGPPRIKV GKKSEHSSEG ELAKLVCKSD ASYPPITDWF WFKTSDTGEE EAITNSTEAN G KYVVVSTP ...String: MAAALLLALA FTLLSGQGAC AAAGTIQTSV QEVNSKTQLT CSLNSSGVDI VGHRWMRGGK VLQEDTLPDL HTKYIVDADD RSGEYSCIF LPEPVGRSEI NVEGPPRIKV GKKSEHSSEG ELAKLVCKSD ASYPPITDWF WFKTSDTGEE EAITNSTEAN G KYVVVSTP EKSQLTISNL DVNVDPGTYV CNATNAQGTT RETISLRVRS RMAALWPFLG IVAEVLVLVT IIFIYEKRRK PD QTLDEDD PGAAPLKGSG THMNDKDKNV RQRNAT UniProtKB: Basigin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: 20 mM HEPES pH 7.5, 150 mM NaCl | |||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 1 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 101.325 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 3.5-4.5 s before plunging. | |||||||||
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 1034 pixel / Digitization - Dimensions - Height: 1034 pixel / Digitization - Frames/image: 1-48 / Number grids imaged: 1 / Number real images: 5895 / Average exposure time: 2.5215 sec. / Average electron dose: 1.56 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | The transmembrane regions of human MCT1 and Basigin-2 were first generated, with side chain of each residue truncated into beta carbon atom. The the model was initial fitted using chimera and then phenix was used for flexible fitting. | ||||||
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Overall B value: 53.6 / Target criteria: Correlation coefficient | ||||||
| Output model |  PDB-7y1q: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)