[English] 日本語
 Yorodumi
Yorodumi- EMDB-33514: Cryo-EM structure of secondary alcohol dehydrogenases TbSADH afte... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of secondary alcohol dehydrogenases TbSADH after carrier-free immobilization based on weak intermolecular interactions | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coordination complex / Activity / Stability / Enzyme Immobilization / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationisopropanol dehydrogenase (NADP+) / isopropanol dehydrogenase (NADP+) activity / zinc ion binding Similarity search - Function | |||||||||
| Biological species |   Thermoanaerobacter brockii (bacteria) Thermoanaerobacter brockii (bacteria) | |||||||||
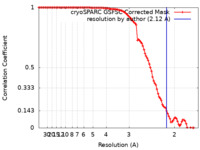
| Method | single particle reconstruction / cryo EM / Resolution: 2.12 Å | |||||||||
 Authors Authors | Chen Q / Li X / Yang F / Qu G / Sun ZT / Wang YJ | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Active and stable alcohol dehydrogenase-assembled hydrogels via synergistic bridging of triazoles and metal ions. Authors: Qiang Chen / Ge Qu / Xu Li / Mingjian Feng / Fan Yang / Yanjie Li / Jincheng Li / Feifei Tong / Shiyi Song / Yujun Wang / Zhoutong Sun / Guangsheng Luo /  Abstract: Biocatalysis is increasingly replacing traditional methods of manufacturing fine chemicals due to its green, mild, and highly selective nature, but biocatalysts, such as enzymes, are generally ...Biocatalysis is increasingly replacing traditional methods of manufacturing fine chemicals due to its green, mild, and highly selective nature, but biocatalysts, such as enzymes, are generally costly, fragile, and difficult to recycle. Immobilization provides protection for the enzyme and enables its convenient reuse, which makes immobilized enzymes promising heterogeneous biocatalysts; however, their industrial applications are limited by the low specific activity and poor stability. Herein, we report a feasible strategy utilizing the synergistic bridging of triazoles and metal ions to induce the formation of porous enzyme-assembled hydrogels with increased activity. The catalytic efficiency of the prepared enzyme-assembled hydrogels toward acetophenone reduction is 6.3 times higher than that of the free enzyme, and the reusability is confirmed by the high residual catalytic activity after 12 cycles of use. A near-atomic resolution (2.1 Å) structure of the hydrogel enzyme is successfully analyzed via cryogenic electron microscopy, which indicates a structure-property relationship for the enhanced performance. In addition, the possible mechanism of gel formation is elucidated, revealing the indispensability of triazoles and metal ions, which guides the use of two other enzymes to prepare enzyme-assembled hydrogels capable of good reusability. The described strategy can pave the way for the development of practical catalytic biomaterials and immobilized biocatalysts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33514.map.gz emd_33514.map.gz | 97 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33514-v30.xml emd-33514-v30.xml emd-33514.xml emd-33514.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33514_fsc.xml emd_33514_fsc.xml | 9.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_33514.png emd_33514.png | 69.3 KB | ||
| Filedesc metadata |  emd-33514.cif.gz emd-33514.cif.gz | 5.6 KB | ||
| Others |  emd_33514_half_map_1.map.gz emd_33514_half_map_1.map.gz emd_33514_half_map_2.map.gz emd_33514_half_map_2.map.gz | 95.3 MB 95.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33514 http://ftp.pdbj.org/pub/emdb/structures/EMD-33514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33514 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33514 | HTTPS FTP |
-Validation report
| Summary document |  emd_33514_validation.pdf.gz emd_33514_validation.pdf.gz | 917.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33514_full_validation.pdf.gz emd_33514_full_validation.pdf.gz | 917.2 KB | Display | |
| Data in XML |  emd_33514_validation.xml.gz emd_33514_validation.xml.gz | 18.2 KB | Display | |
| Data in CIF |  emd_33514_validation.cif.gz emd_33514_validation.cif.gz | 22.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33514 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33514 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33514 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33514 | HTTPS FTP |
-Related structure data
| Related structure data |  7xy9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33514.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33514.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8374 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33514_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
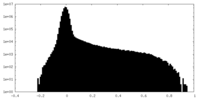
| Projections & Slices |
| ||||||||||||
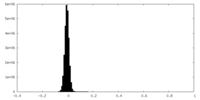
| Density Histograms |
-Half map: #1
| File | emd_33514_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
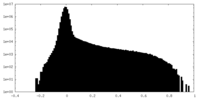
| Projections & Slices |
| ||||||||||||
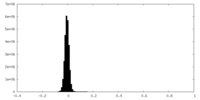
| Density Histograms |
- Sample components
Sample components
-Entire : enzyme assembled gel
| Entire | Name: enzyme assembled gel |
|---|---|
| Components |
|
-Supramolecule #1: enzyme assembled gel
| Supramolecule | Name: enzyme assembled gel / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Thermoanaerobacter brockii (bacteria) Thermoanaerobacter brockii (bacteria) |
-Macromolecule #1: NADP-dependent isopropanol dehydrogenase
| Macromolecule | Name: NADP-dependent isopropanol dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: isopropanol dehydrogenase (NADP+) |
|---|---|
| Source (natural) | Organism:   Thermoanaerobacter brockii (bacteria) Thermoanaerobacter brockii (bacteria) |
| Molecular weight | Theoretical: 38.524695 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHKGF AMLSIGKVGW IEKEKPAPGP FDAIVRPLAV APCTSDIHTV FEGAIGERHN MILGHEAVGE VVEVGSEVKD FKPGDRVVV PANTPDWRTS EVQRGYHQHS GGMLAGWKFS NVKDGVFGEF FHVNDADMNL AHLPKEIPLE AAVMIPDMMT T GFHGAELA ...String: MHHHHHHKGF AMLSIGKVGW IEKEKPAPGP FDAIVRPLAV APCTSDIHTV FEGAIGERHN MILGHEAVGE VVEVGSEVKD FKPGDRVVV PANTPDWRTS EVQRGYHQHS GGMLAGWKFS NVKDGVFGEF FHVNDADMNL AHLPKEIPLE AAVMIPDMMT T GFHGAELA DIELGATVAV LGIGPVGLMA VAGAKLRGAG RIIAVGSRPV CVDAAKYYGA TDIVNYKDGP IESQIMNLTE GK GVDAAII AGGNADIMAT AVKIVKPGGT IANVNYFGEG EVLPVPRLEW GCGMAHKTIK GGLCPGGRLR MERLIDLVFY KRV DPSKLV THVFRGFDNI EKAFMLMKDK PKDLIKPVVI LA UniProtKB: NADP-dependent isopropanol dehydrogenase |
-Macromolecule #2: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 2 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 8 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 574 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)