[English] 日本語
 Yorodumi
Yorodumi- EMDB-33504: Cryo-EM structure of the purinergic receptor P2Y12R in complex wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the purinergic receptor P2Y12R in complex with 2MeSADP and Gi | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | G protein-coupled receptor / purinergic receptor / P2Y12R / Ligand binding / signal transduction / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationvisual system development / positive regulation of integrin activation by cell surface receptor linked signal transduction / regulation of microglial cell migration / G protein-coupled ADP receptor activity / cerebral cortex radial glia-guided migration / cell body membrane / P2Y receptors / G protein-coupled purinergic nucleotide receptor activity / positive regulation of microglial cell migration / positive regulation of monoatomic ion transport ...visual system development / positive regulation of integrin activation by cell surface receptor linked signal transduction / regulation of microglial cell migration / G protein-coupled ADP receptor activity / cerebral cortex radial glia-guided migration / cell body membrane / P2Y receptors / G protein-coupled purinergic nucleotide receptor activity / positive regulation of microglial cell migration / positive regulation of monoatomic ion transport / negative regulation of adenylate cyclase-activating adrenergic receptor signaling pathway / G protein-coupled adenosine receptor activity / hemostasis / cell projection membrane / negative regulation of calcium ion-dependent exocytosis / substrate-dependent cell migration, cell extension / G protein-coupled adenosine receptor signaling pathway / regulation of chemotaxis / positive regulation of chemotaxis / negative regulation of adenylate cyclase activity / positive regulation of urine volume / cell projection organization / positive regulation of neural precursor cell proliferation / negative regulation of synaptic transmission / positive regulation of cell adhesion mediated by integrin / positive regulation of ruffle assembly / lamellipodium assembly / gamma-aminobutyric acid signaling pathway / cellular response to ATP / regulation of calcium ion transport / negative regulation of apoptotic signaling pathway / neuronal dense core vesicle / response to axon injury / positive regulation of vascular associated smooth muscle cell proliferation / positive regulation of superoxide anion generation / Adenylate cyclase inhibitory pathway / monoatomic ion transport / response to nutrient / guanyl-nucleotide exchange factor activity / hippocampal mossy fiber to CA3 synapse / Regulation of insulin secretion / establishment of localization in cell / calcium-mediated signaling / G protein-coupled receptor binding / platelet activation / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / platelet aggregation / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / cell body / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||||||||
 Authors Authors | Tan Q / Li B / Han S / Zhao Q / Wu B | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2023 Journal: Protein Cell / Year: 2023Title: Structural insights into signal transduction of the purinergic receptors P2Y1R and P2Y12R. Authors: Beibei Li / Shuo Han / Mu Wang / Yu Yu / Limin Ma / Xiaojing Chu / Qiuxiang Tan / Qiang Zhao / Beili Wu /  | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33504.map.gz emd_33504.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33504-v30.xml emd-33504-v30.xml emd-33504.xml emd-33504.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33504.png emd_33504.png | 40.3 KB | ||
| Filedesc metadata |  emd-33504.cif.gz emd-33504.cif.gz | 7.1 KB | ||
| Others |  emd_33504_half_map_1.map.gz emd_33504_half_map_1.map.gz emd_33504_half_map_2.map.gz emd_33504_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33504 http://ftp.pdbj.org/pub/emdb/structures/EMD-33504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33504 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33504 | HTTPS FTP |
-Validation report
| Summary document |  emd_33504_validation.pdf.gz emd_33504_validation.pdf.gz | 843.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33504_full_validation.pdf.gz emd_33504_full_validation.pdf.gz | 843.1 KB | Display | |
| Data in XML |  emd_33504_validation.xml.gz emd_33504_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_33504_validation.cif.gz emd_33504_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33504 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33504 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33504 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33504 | HTTPS FTP |
-Related structure data
| Related structure data |  7xxiMC  7xxhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33504.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33504.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33504_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33504_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The purinergic receptor P2Y12R in complex with 2MeSADP and Gi
| Entire | Name: The purinergic receptor P2Y12R in complex with 2MeSADP and Gi |
|---|---|
| Components |
|
-Supramolecule #1: The purinergic receptor P2Y12R in complex with 2MeSADP and Gi
| Supramolecule | Name: The purinergic receptor P2Y12R in complex with 2MeSADP and Gi type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: P2Y purinoceptor 12
| Macromolecule | Name: P2Y purinoceptor 12 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 43.665078 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAPQAVDNLT SAPGNTSLCT RDYKITQVLF PLLYTVLFFV GLITNGLAMR IFFQIRSKSN FIIFLKNTVI SDLLMILTFP FKILSDAKL GTGPLRTFVC QVTSVIFYFT MYISISFLGL ITIDRYQKTT RPFKTSNPKN LLGAKILSVV IWAFMFLLSL P NMILTNRQ ...String: GAPQAVDNLT SAPGNTSLCT RDYKITQVLF PLLYTVLFFV GLITNGLAMR IFFQIRSKSN FIIFLKNTVI SDLLMILTFP FKILSDAKL GTGPLRTFVC QVTSVIFYFT MYISISFLGL ITIDRYQKTT RPFKTSNPKN LLGAKILSVV IWAFMFLLSL P NMILTNRQ PRDKNVKKCS FLKSEFGLVW HEIVNYICQV IFWINFLIVI VCYTLITKEL YRSYVRTRGV GKVPRKKVNV KV FIIIAVF FICFVPFHFA RIPYTLSQTR DVFDCTAENT LFYVKESTLW LTSLNACLDP FIYFFLCKSF RNSLISMLKC PNS ATSLSQ DNRKKEQDGG DPNEETPMEF LEVLFQGPGS WSHPQFEKGS GAGASAGSWS HPQFEK UniProtKB: P2Y purinoceptor 12 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-2 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.502863 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTVSAEDK AAAERSKMID KNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEDG YSEEECRQYR AVVYSNTIQS IMAIVKAMG NLQIDFADPS RADDARQLFA LSCTAEEQGV LPDDLSGVIR RLWADHGVQA CFGRSREYQL NDSAAYYLND L ERIAQSDY ...String: MGCTVSAEDK AAAERSKMID KNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEDG YSEEECRQYR AVVYSNTIQS IMAIVKAMG NLQIDFADPS RADDARQLFA LSCTAEEQGV LPDDLSGVIR RLWADHGVQA CFGRSREYQL NDSAAYYLND L ERIAQSDY IPTQQDVLRT RVKTTGIVET HFTFKDLHFK MFDVGAQRSE RKKWIHCFEG VTAIIFCVAL SAYDLVLAED EE MNRMHAS MKLFDSICNN KWFTDTSIIL FLNKKDLFEE KITHSPLTIC FPEYTGANKY DEAASYIQSK FEDLNKRKDT KEI YTHFTC STDTKNVQFV FDAVTDVIIK NNLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-2 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.245805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD D NQIVTSSG ...String: MHHHHHHSEL DQLRQEAEQL KNQIRDARKA CADATLSQIT NNIDPVGRIQ MRTRRTLRGH LAKIYAMHWG TDSRLLVSAS QDGKLIIWD SYTTNKVHAI PLRSSWVMTC AYAPSGNYVA CGGLDNICSI YNLKTREGNV RVSRELAGHT GYLSCCRFLD D NQIVTSSG DTTCALWDIE TGQQTTTFTG HTGDVMSLSL APDTRLFVSG ACDASAKLWD VREGMCRQTF TGHESDINAI CF FPNGNAF ATGSDDATCR LFDLRADQEL MTYSHDNIIC GITSVSFSKS GRLLLAGYDD FNCNVWDALK ADRAGVLAGH DNR VSCLGV TDDGMAVATG SWDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate)
| Macromolecule | Name: 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate) type: ligand / ID: 5 / Number of copies: 1 / Formula: 6AD |
|---|---|
| Molecular weight | Theoretical: 473.293 Da |
| Chemical component information | 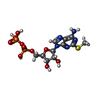 ChemComp-6AD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)