+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | IAA bound state of AtPIN3 | |||||||||
 Map data Map data | IAA bound state of AtPIN3 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | PIN-FORMED (PIN) protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtropism / root hair initiation / root hair elongation / gravitropism / positive gravitropism / auxin export across the plasma membrane / auxin efflux transmembrane transporter activity / auxin polar transport / root development / auxin-activated signaling pathway ...tropism / root hair initiation / root hair elongation / gravitropism / positive gravitropism / auxin export across the plasma membrane / auxin efflux transmembrane transporter activity / auxin polar transport / root development / auxin-activated signaling pathway / vesicle membrane / response to light stimulus / lateral plasma membrane / cell surface / protein homodimerization activity / mitochondrion / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.93 Å | |||||||||
 Authors Authors | Su N | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structures and mechanisms of the Arabidopsis auxin transporter PIN3. Authors: Nannan Su / Aiqin Zhu / Xin Tao / Zhong Jie Ding / Shenghai Chang / Fan Ye / Yan Zhang / Cheng Zhao / Qian Chen / Jiangqin Wang / Chen Yu Zhou / Yirong Guo / Shasha Jiao / Sufen Zhang / Han ...Authors: Nannan Su / Aiqin Zhu / Xin Tao / Zhong Jie Ding / Shenghai Chang / Fan Ye / Yan Zhang / Cheng Zhao / Qian Chen / Jiangqin Wang / Chen Yu Zhou / Yirong Guo / Shasha Jiao / Sufen Zhang / Han Wen / Lixin Ma / Sheng Ye / Shao Jian Zheng / Fan Yang / Shan Wu / Jiangtao Guo /  Abstract: The PIN-FORMED (PIN) protein family of auxin transporters mediates polar auxin transport and has crucial roles in plant growth and development. Here we present cryo-electron microscopy structures of ...The PIN-FORMED (PIN) protein family of auxin transporters mediates polar auxin transport and has crucial roles in plant growth and development. Here we present cryo-electron microscopy structures of PIN3 from Arabidopsis thaliana in the apo state and in complex with its substrate indole-3-acetic acid and the inhibitor N-1-naphthylphthalamic acid (NPA). A. thaliana PIN3 exists as a homodimer, and its transmembrane helices 1, 2 and 7 in the scaffold domain are involved in dimerization. The dimeric PIN3 forms a large, joint extracellular-facing cavity at the dimer interface while each subunit adopts an inward-facing conformation. The structural and functional analyses, along with computational studies, reveal the structural basis for the recognition of indole-3-acetic acid and NPA and elucidate the molecular mechanism of NPA inhibition on PIN-mediated auxin transport. The PIN3 structures support an elevator-like model for the transport of auxin, whereby the transport domains undergo up-down rigid-body motions and the dimerized scaffold domains remain static. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33500.map.gz emd_33500.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33500-v30.xml emd-33500-v30.xml emd-33500.xml emd-33500.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33500.png emd_33500.png | 50.3 KB | ||
| Masks |  emd_33500_msk_1.map emd_33500_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33500.cif.gz emd-33500.cif.gz | 6.6 KB | ||
| Others |  emd_33500_half_map_1.map.gz emd_33500_half_map_1.map.gz emd_33500_half_map_2.map.gz emd_33500_half_map_2.map.gz | 39.7 MB 39.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33500 http://ftp.pdbj.org/pub/emdb/structures/EMD-33500 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33500 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33500 | HTTPS FTP |
-Related structure data
| Related structure data |  7xxbMC  7wksC  7wkwC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33500.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33500.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | IAA bound state of AtPIN3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.93 Å | ||||||||||||||||||||||||||||||||||||
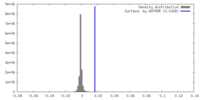
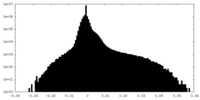
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33500_msk_1.map emd_33500_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
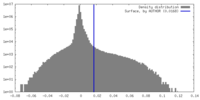
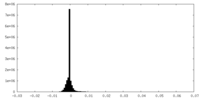
| Density Histograms |
-Half map: half1 map of AtPIN3 IAA
| File | emd_33500_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 map of AtPIN3_IAA | ||||||||||||
| Projections & Slices |
| ||||||||||||
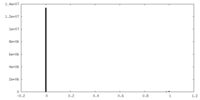
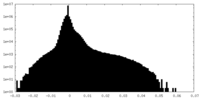
| Density Histograms |
-Half map: half2 map of AtPIN3 IAA
| File | emd_33500_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 map of AtPIN3_IAA | ||||||||||||
| Projections & Slices |
| ||||||||||||
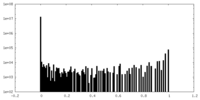
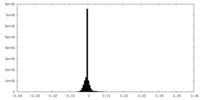
| Density Histograms |
- Sample components
Sample components
-Entire : IAA-bound state AtPIN3(AtPIN3-IAA)
| Entire | Name: IAA-bound state AtPIN3(AtPIN3-IAA) |
|---|---|
| Components |
|
-Supramolecule #1: IAA-bound state AtPIN3(AtPIN3-IAA)
| Supramolecule | Name: IAA-bound state AtPIN3(AtPIN3-IAA) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Auxin efflux carrier component 3
| Macromolecule | Name: Auxin efflux carrier component 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 73.983531 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DYKDDDDKWS HPQFEKGGGG SGGSAWSHPQ FEKEFKGLVD MISWHDLYTV LTAVIPLYVA MILAYGSVRW WKIFSPDQCS GINRFVAIF AVPLLSFHFI STNNPYAMNL RFIAADTLQK IIMLSLLVLW ANFTRSGSLE WSITIFSLST LPNTLVMGIP L LIAMYGEY ...String: DYKDDDDKWS HPQFEKGGGG SGGSAWSHPQ FEKEFKGLVD MISWHDLYTV LTAVIPLYVA MILAYGSVRW WKIFSPDQCS GINRFVAIF AVPLLSFHFI STNNPYAMNL RFIAADTLQK IIMLSLLVLW ANFTRSGSLE WSITIFSLST LPNTLVMGIP L LIAMYGEY SGSLMVQIVV LQCIIWYTLL LFLFEFRGAK MLIMEQFPET AASIVSFKVE SDVVSLDGHD FLETDAEIGD DG KLHVTVR KSNASRRSFC GPNMTPRPSN LTGAEIYSLS TTPRGSNFNH SDFYNMMGFP GGRLSNFGPA DMYSVQSSRG PTP RPSNFE ENCAMASSPR FGYYPGGGAG SYPAPNPEFS STTTSTANKS VNKNPKDVNT NQQTTLPTGG KSNSHDAKEL HMFV WSSNG SPVSDRAGLN VFGGAPDNDQ GGRSDQGAKE IRMLVPDQSH NGETKAVAHP ASGDFGGEQQ FSFAGKEEEA ERPKD AENG LNKLAPNSTA ALQSKTGLGG AEASQRKNMP PASVMTRLIL IMVWRKLIRN PNTYSSLIGL IWALVAFRWH VAMPKI IQQ SISILSDAGL GMAMFSLGLF MALQPKLIAC GNSVATFAMA VRFLTGPAVM AVAAIAIGLR GDLLRVAIVQ AALPQGI VP FVFAKEYNVH PAILSTGVIF GMLIALPITL VYYILLGL UniProtKB: Auxin efflux carrier component 3 |
-Macromolecule #2: 1H-INDOL-3-YLACETIC ACID
| Macromolecule | Name: 1H-INDOL-3-YLACETIC ACID / type: ligand / ID: 2 / Number of copies: 2 / Formula: IAC |
|---|---|
| Molecular weight | Theoretical: 175.184 Da |
| Chemical component information | 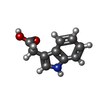 ChemComp-IAC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average exposure time: 6.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)