+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human SLC26A3 in complex with UK5099 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibitor / Transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC26A3 causes congenital secretory chloride diarrhea 1 (DIAR1) / Inorganic anion exchange by SLC26 transporters / sulfate transmembrane transporter activity / oxalate transmembrane transporter activity / sulfate transmembrane transport / secondary active sulfate transmembrane transporter activity / intracellular pH elevation / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity ...Defective SLC26A3 causes congenital secretory chloride diarrhea 1 (DIAR1) / Inorganic anion exchange by SLC26 transporters / sulfate transmembrane transporter activity / oxalate transmembrane transporter activity / sulfate transmembrane transport / secondary active sulfate transmembrane transporter activity / intracellular pH elevation / chloride:bicarbonate antiporter activity / solute:inorganic anion antiporter activity / bicarbonate transmembrane transporter activity / monoatomic anion transport / membrane hyperpolarization / chloride transmembrane transporter activity / sperm capacitation / monoatomic ion transport / chloride transmembrane transport / cellular response to cAMP / brush border membrane / sperm midpiece / apical plasma membrane / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Li XR / Chi XM / Zhang YY / Zhou Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: human SLC26A3 in complex with UK5099 Authors: Li XR / Chi XM / Zhang YY / Zhou Q | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33471.map.gz emd_33471.map.gz | 35.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33471-v30.xml emd-33471-v30.xml emd-33471.xml emd-33471.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33471.png emd_33471.png | 49.9 KB | ||
| Filedesc metadata |  emd-33471.cif.gz emd-33471.cif.gz | 6.1 KB | ||
| Others |  emd_33471_half_map_1.map.gz emd_33471_half_map_1.map.gz emd_33471_half_map_2.map.gz emd_33471_half_map_2.map.gz | 28.1 MB 28.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33471 http://ftp.pdbj.org/pub/emdb/structures/EMD-33471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33471 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33471 | HTTPS FTP |
-Related structure data
| Related structure data |  7xujMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33471.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33471.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.077 Å | ||||||||||||||||||||||||||||||||||||
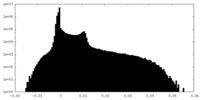
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33471_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
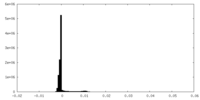
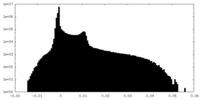
| Density Histograms |
-Half map: #1
| File | emd_33471_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
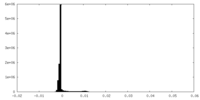
| Density Histograms |
- Sample components
Sample components
-Entire : human SLC26A3 in complex with UK5099
| Entire | Name: human SLC26A3 in complex with UK5099 |
|---|---|
| Components |
|
-Supramolecule #1: human SLC26A3 in complex with UK5099
| Supramolecule | Name: human SLC26A3 in complex with UK5099 / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Chloride anion exchanger
| Macromolecule | Name: Chloride anion exchanger / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 79.333672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QYIVARPVYS TNAFEENHKK TGRHHKTFLD HLKVCCSCSP QKAKRIVLSL FPIASWLPAY RLKEWLLSDI VSGISTGIVA VLQGLAFAL LVDIPPVYGL YASFFPAIIY LFFGTSRHIS VGPFPILSMM VGLAVSGAVS KAVPDRNATT LGLPNNSNNS S LLDDERVR ...String: QYIVARPVYS TNAFEENHKK TGRHHKTFLD HLKVCCSCSP QKAKRIVLSL FPIASWLPAY RLKEWLLSDI VSGISTGIVA VLQGLAFAL LVDIPPVYGL YASFFPAIIY LFFGTSRHIS VGPFPILSMM VGLAVSGAVS KAVPDRNATT LGLPNNSNNS S LLDDERVR VAAAASVTVL SGIIQLAFGI LRIGFVVIYL SESLISGFTT AAAVHVLVSQ LKFIFQLTVP SHTDPVSIFK VL YSVFSQI EKTNIADLVT ALIVLLVVSI VKEINQRFKD KLPVPIPIEF IMTVIAAGVS YGCDFKNRFK VAVVGDMNPG FQP PITPDV ETFQNTVGDC FGIAMVAFAV AFSVASVYSL KYDYPLDGNQ ELIALGLGNI VCGVFRGFAG STALSRSAVQ ESTG GKTQI AGLIGAIIVL IVVLAIGFLL APLQKSVLAA LALGNLKGML MQFAEIGRLW RKDKYDCLIW IMTFIFTIVL GLGLG LAAS VAFQLLTIVF RTQFPKCSTL ANIGRTNIYK NKKDYYDMYE PEGVKIFRCP SPIYFANIGF FRRKLIDAVG FSPLRI LRK RNKALRKIRK LQKQGLLQVT PKGFICTVDT IKDSDEELDN NQIEVLDQPI NTTDLPFHID WNDDLPLNIE VPKISLH SL ILDFSAVSFL DVSSVRGLKS ILQEFIRIKV DVYIVGTDDD FIEKLNRYEF FDGEVKSSIF FLTIHDAVLH ILMKKD UniProtKB: Chloride anion exchanger |
-Macromolecule #2: (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid
| Macromolecule | Name: (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid / type: ligand / ID: 2 / Number of copies: 2 / Formula: I2R |
|---|---|
| Molecular weight | Theoretical: 288.3 Da |
| Chemical component information | 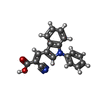 ChemComp-I2R: |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 8 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)