[English] 日本語
 Yorodumi
Yorodumi- EMDB-33434: Cryo-EM structure of SARS-CoV-2 Omicron spike glycoprotein in com... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 Omicron spike glycoprotein in complex with three F61 Fab and three D2 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Omicron spike / antibody / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
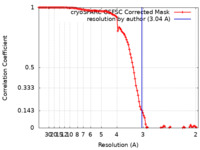
| Method | single particle reconstruction / cryo EM / Resolution: 3.04 Å | |||||||||
 Authors Authors | Wang X / Li X | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2022 Journal: Cell Discov / Year: 2022Title: Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages. Authors: Xiaoman Li / Yongbing Pan / Qiangling Yin / Zejun Wang / Sisi Shan / Laixing Zhang / Jinfang Yu / Yuanyuan Qu / Lina Sun / Fang Gui / Jia Lu / Zhaofei Jing / Wei Wu / Tao Huang / Xuanling ...Authors: Xiaoman Li / Yongbing Pan / Qiangling Yin / Zejun Wang / Sisi Shan / Laixing Zhang / Jinfang Yu / Yuanyuan Qu / Lina Sun / Fang Gui / Jia Lu / Zhaofei Jing / Wei Wu / Tao Huang / Xuanling Shi / Jiandong Li / Xinguo Li / Dexin Li / Shiwen Wang / Maojun Yang / Linqi Zhang / Kai Duan / Mifang Liang / Xiaoming Yang / Xinquan Wang /  Abstract: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs), especially the latest Omicron, have exhibited severe antibody evasion. Broadly neutralizing antibodies ...The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs), especially the latest Omicron, have exhibited severe antibody evasion. Broadly neutralizing antibodies with high potency against Omicron are urgently needed for understanding the working mechanisms and developing therapeutic agents. In this study, we characterized the previously reported F61, which was isolated from convalescent patients infected with prototype SARS-CoV-2, as a broadly neutralizing antibody against all VOCs including Omicron BA.1, BA.1.1, BA.2, BA.3 and BA.4 sublineages by utilizing antigen binding and cell infection assays. We also identified and characterized another broadly neutralizing antibody D2 with epitope distinct from that of F61. More importantly, we showed that a combination of F61 with D2 exhibited synergy in neutralization and protecting mice from SARS-CoV-2 Delta and Omicron BA.1 variants. Cryo-Electron Microscopy (Cryo-EM) structures of the spike-F61 and spike-D2 binary complexes revealed the distinct epitopes of F61 and D2 at atomic level and the structural basis for neutralization. Cryo-EM structure of the Omicron-spike-F61-D2 ternary complex provides further structural insights into the synergy between F61 and D2. These results collectively indicated F61 and F61-D2 cocktail as promising therapeutic antibodies for combating SARS-CoV-2 variants including diverse Omicron sublineages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33434.map.gz emd_33434.map.gz | 141.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33434-v30.xml emd-33434-v30.xml emd-33434.xml emd-33434.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33434_fsc.xml emd_33434_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33434.png emd_33434.png | 49.4 KB | ||
| Filedesc metadata |  emd-33434.cif.gz emd-33434.cif.gz | 6.6 KB | ||
| Others |  emd_33434_additional_1.map.gz emd_33434_additional_1.map.gz emd_33434_half_map_1.map.gz emd_33434_half_map_1.map.gz emd_33434_half_map_2.map.gz emd_33434_half_map_2.map.gz | 141.5 MB 139.2 MB 139.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33434 http://ftp.pdbj.org/pub/emdb/structures/EMD-33434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33434 | HTTPS FTP |
-Validation report
| Summary document |  emd_33434_validation.pdf.gz emd_33434_validation.pdf.gz | 815.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33434_full_validation.pdf.gz emd_33434_full_validation.pdf.gz | 815.4 KB | Display | |
| Data in XML |  emd_33434_validation.xml.gz emd_33434_validation.xml.gz | 19.7 KB | Display | |
| Data in CIF |  emd_33434_validation.cif.gz emd_33434_validation.cif.gz | 25.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33434 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33434 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33434 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33434 | HTTPS FTP |
-Related structure data
| Related structure data |  7xstMC  7xmxC  7xmzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33434.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33434.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
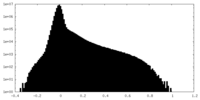
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: local map
| File | emd_33434_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local map | ||||||||||||
| Projections & Slices |
| ||||||||||||
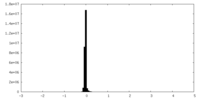
| Density Histograms |
-Half map: #2
| File | emd_33434_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
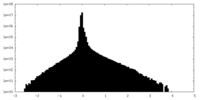
| Density Histograms |
-Half map: #1
| File | emd_33434_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
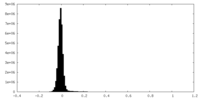
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Omicron spike in complex with F61 and D2 Fab
| Entire | Name: SARS-CoV-2 Omicron spike in complex with F61 and D2 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Omicron spike in complex with F61 and D2 Fab
| Supramolecule | Name: SARS-CoV-2 Omicron spike in complex with F61 and D2 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.628844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ PFLMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPIIVR EPEDLPQGFS ALEPLVDLPI GINITRFQTL LA LHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFD EVFNATRFAS VYAWNRKRIS NCVADYSVLY NLAPFFTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKVSGN YNYLYRLFRK SNLKPFERDI STEIYQAGNK PCNGV AGFN CYFPLRSYSF RPTYGVGHQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLK GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEY VNNSYECDIP IGAGICASYQ TQTKSHGSAS SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLK RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KYFGGFNFS QILPDPSKPS KRSPIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFKGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGP ALQIPFPMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTP SALGKLQDVV NHNAQALNTL V KQLSSKFG AISSVLNDIF SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ YIKWP UniProtKB: Spike glycoprotein |
-Macromolecule #2: F61 heavy chain
| Macromolecule | Name: F61 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 12.993491 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCEASEIIVN RNYMNWVRQA PGKGLEWVSI IYPGGSTFYA DSVKGRFTIS RDNSKNTMYL QMNSLRAED TAVYYCARSY GDFYVDFWGQ GTLVTVSS |
-Macromolecule #3: F61 light chain
| Macromolecule | Name: F61 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.472622 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SAAPGQKVTI SCSGSSSNIG NNYVSWYQQL PGTAPKLLIY DNNKRPSGIP DRFSGSKSGT SATLGITGLQ TGDEADYYC GTWDSSLSAW VFGGGTKLTV L |
-Macromolecule #4: D2 heavy chain
| Macromolecule | Name: D2 heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.406785 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGGG LVQPGRSLTL SCGASGFTFE DYAMHWVRQA PGKGLEWVSG IDWNSGVIGY ADSVKGRFII SRDNAKNSLY LHMRSLTAE DTALYYCAKD VYSESGSGSY YDYWGQGTLV TVSS |
-Macromolecule #5: D2 light chain
| Macromolecule | Name: D2 light chain / type: protein_or_peptide / ID: 5 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.468719 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSVLTQPPSV SAAPGQKVAI SCSGSTSNIG DNFVSWYQQF PGTAPKLLLY DDARRPSGIP DRFSGSKSGT SATLGITGLQ TGDEAVYFC STWDNSLNVV LFGGGTKLTV L |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 31 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.91 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)