+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human B-cell antigen receptor of the IgM isotype | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | immunology / B cell signalling / adaptive immunity / antibody / SIGNALING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationIgM B cell receptor complex / IgD immunoglobulin complex / B cell receptor complex / IgA immunoglobulin complex / IgM immunoglobulin complex / IgE immunoglobulin complex / CD22 mediated BCR regulation / IgG immunoglobulin complex / Fc epsilon receptor (FCERI) signaling / Classical antibody-mediated complement activation ...IgM B cell receptor complex / IgD immunoglobulin complex / B cell receptor complex / IgA immunoglobulin complex / IgM immunoglobulin complex / IgE immunoglobulin complex / CD22 mediated BCR regulation / IgG immunoglobulin complex / Fc epsilon receptor (FCERI) signaling / Classical antibody-mediated complement activation / Initial triggering of complement / immunoglobulin mediated immune response / B cell activation / B cell proliferation / FCGR activation / Role of LAT2/NTAL/LAB on calcium mobilization / Role of phospholipids in phagocytosis / Scavenging of heme from plasma / antigen binding / multivesicular body / FCERI mediated Ca+2 mobilization / FCGR3A-mediated IL10 synthesis / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / B cell differentiation / Regulation of Complement cascade / Cell surface interactions at the vascular wall / B cell receptor signaling pathway / FCGR3A-mediated phagocytosis / FCERI mediated MAPK activation / response to bacterium / Regulation of actin dynamics for phagocytic cup formation / FCERI mediated NF-kB activation / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / transmembrane signaling receptor activity / blood microparticle / Potential therapeutics for SARS / adaptive immune response / immune response / membrane raft / external side of plasma membrane / signal transduction / extracellular space / extracellular exosome / extracellular region / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||
 Authors Authors | Chen MY / Su Q / Shi YG | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Cryo-EM structure of the human IgM B cell receptor. Authors: Qiang Su / Mengying Chen / Yan Shi / Xiaofeng Zhang / Gaoxingyu Huang / Bangdong Huang / Dongwei Liu / Zhangsuo Liu / Yigong Shi /  Abstract: The B cell receptor (BCR) initiates immune responses through antigen recognition. We report a 3.3-angstrom cryo-electron microscopy structure of human immunoglobulin M (IgM)-BCR in the resting state. ...The B cell receptor (BCR) initiates immune responses through antigen recognition. We report a 3.3-angstrom cryo-electron microscopy structure of human immunoglobulin M (IgM)-BCR in the resting state. IgM-BCR comprises two heavy chains, two light chains, and the Igα/Igβ heterodimer. The ectodomains of the heavy chains closely stack against those of Igα/Igβ, with one heavy chain locked between Igα and Igβ in the juxtamembrane region. Extracellular interactions may determine isotype specificity of the BCR. The transmembrane helices of IgM-BCR form a four-helix bundle that appears to be conserved among all BCR isotypes. This structure contains 14 glycosylation sites on the IgM-BCR ectodomains and reveals three potential surface binding sites. Our work reveals the organizational principles of the BCR and may facilitate the design of antibody-based therapeutics. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33390.map.gz emd_33390.map.gz | 167.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33390-v30.xml emd-33390-v30.xml emd-33390.xml emd-33390.xml | 23.8 KB 23.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33390.png emd_33390.png | 22.1 KB | ||
| Filedesc metadata |  emd-33390.cif.gz emd-33390.cif.gz | 7.1 KB | ||
| Others |  emd_33390_half_map_1.map.gz emd_33390_half_map_1.map.gz emd_33390_half_map_2.map.gz emd_33390_half_map_2.map.gz | 164.7 MB 164.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33390 http://ftp.pdbj.org/pub/emdb/structures/EMD-33390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33390 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33390 | HTTPS FTP |
-Related structure data
| Related structure data |  7xq8MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33390.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33390.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.077 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33390_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
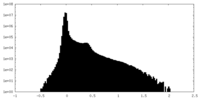
| Projections & Slices |
| ||||||||||||
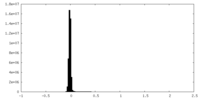
| Density Histograms |
-Half map: #1
| File | emd_33390_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
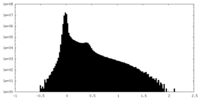
| Projections & Slices |
| ||||||||||||
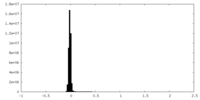
| Density Histograms |
- Sample components
Sample components
-Entire : structure of human B-cell antigen receptor of the IgM isotype
| Entire | Name: structure of human B-cell antigen receptor of the IgM isotype |
|---|---|
| Components |
|
-Supramolecule #1: structure of human B-cell antigen receptor of the IgM isotype
| Supramolecule | Name: structure of human B-cell antigen receptor of the IgM isotype type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Heavy chain
| Supramolecule | Name: Heavy chain / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Light chain
| Supramolecule | Name: Light chain / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #4: human B-cell antigen receptor
| Supramolecule | Name: human B-cell antigen receptor / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3-#4 |
|---|
-Macromolecule #1: Chimera of Heavy chain of VRC01 antibody Fab and Isoform 2 of Imm...
| Macromolecule | Name: Chimera of Heavy chain of VRC01 antibody Fab and Isoform 2 of Immunoglobulin heavy constant mu type: protein_or_peptide / ID: 1 Details: Specific N-terminal secretion signal peptide (1-19) Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.895641 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEFGLSWLFL VAILKGVQCQ VQLVQSGGQM KKPGESMRIS CRASGYEFID CTLNWIRLAP GKRPEWMGWL KPRGGAVNYA RPLQGRVTM TRDVYSDTAF LELRSLTVDD TAVYFCTRGK NCDYNWDFEH WGRGTPVIVS SGSASAPTLF PLVSCENSPS D TSSVAVGC ...String: MEFGLSWLFL VAILKGVQCQ VQLVQSGGQM KKPGESMRIS CRASGYEFID CTLNWIRLAP GKRPEWMGWL KPRGGAVNYA RPLQGRVTM TRDVYSDTAF LELRSLTVDD TAVYFCTRGK NCDYNWDFEH WGRGTPVIVS SGSASAPTLF PLVSCENSPS D TSSVAVGC LAQDFLPDSI TFSWKYKNNS DISSTRGFPS VLRGGKYAAT SQVLLPSKDV MQGTDEHVVC KVQHPNGNKE KN VPLPVIA ELPPKVSVFV PPRDGFFGNP RKSKLICQAT GFSPRQIQVS WLREGKQVGS GVTTDQVQAE AKESGPTTYK VTS TLTIKE SDWLGQSMFT CRVDHRGLTF QQNASSMCVP DQDTAIRVFA IPPSFASIFL TKSTKLTCLV TDLTTYDSVT ISWT RQNGE AVKTHTNISE SHPNATFSAV GEASICEDDW NSGERFTCTV THTDLPSPLK QTISRPKGVA LHRPDVYLLP PAREQ LNLR ESATITCLVT GFSPADVFVQ WMQRGQPLSP EKYVTSAPMP EPQAPGRYFA HSILTVSEEE WNTGETYTCV VAHEAL PNR VTERTVDKST EGEVSADEEG FENLWATAST FIVLFLLSLF YSTTVTLFKV K UniProtKB: Isoform 2 of Immunoglobulin heavy constant mu |
-Macromolecule #2: Light chain of Fab fragments of the VRC01 antibody,Immunoglobulin...
| Macromolecule | Name: Light chain of Fab fragments of the VRC01 antibody,Immunoglobulin kappa constant type: protein_or_peptide / ID: 2 Details: N terminal specific signal peptide (1-19), Flag tag (20-40) Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.350191 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVLQTQVFIS LLLWISGAYG GSDYKDDDDK GSPGDEVDAG EIVLTQSPGT LSLSPGETAI ISCRTSQYGS LAWYQQRPGQ APRLVIYSG STRAAGIPDR FSGSRWGPDY NLTISNLESG DFGVYYCQQY EFFGQGTKVQ VDIKRTVAAP SVFIFPPSDE Q LKSGTASV ...String: MVLQTQVFIS LLLWISGAYG GSDYKDDDDK GSPGDEVDAG EIVLTQSPGT LSLSPGETAI ISCRTSQYGS LAWYQQRPGQ APRLVIYSG STRAAGIPDR FSGSRWGPDY NLTISNLESG DFGVYYCQQY EFFGQGTKVQ VDIKRTVAAP SVFIFPPSDE Q LKSGTASV VCLLNNFYPR EAKVQWKVDN ALQSGNSQES VTEQDSKDST YSLSSTLTLS KADYEKHKVY ACEVTHQGLS SP VTKSFNR GEC UniProtKB: Immunoglobulin kappa constant |
-Macromolecule #3: B-cell antigen receptor complex-associated protein alpha chain
| Macromolecule | Name: B-cell antigen receptor complex-associated protein alpha chain type: protein_or_peptide / ID: 3 / Details: C terminal twin-strep tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.144594 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPGGPGVLQA LPATIFLLFL LSAVYLGPGC QALWMHKVPA SLMVSLGEDA HFQCPHNSSN NANVTWWRVL HGNYTWPPEF LGPGEDPNG TLIIQNVNKS HGGIYVCRVQ EGNESYQQSC GTYLRVRQPP PRPFLDMGEG TKNRIITAEG IILLFCAVVP G TLLLFRKR ...String: MPGGPGVLQA LPATIFLLFL LSAVYLGPGC QALWMHKVPA SLMVSLGEDA HFQCPHNSSN NANVTWWRVL HGNYTWPPEF LGPGEDPNG TLIIQNVNKS HGGIYVCRVQ EGNESYQQSC GTYLRVRQPP PRPFLDMGEG TKNRIITAEG IILLFCAVVP G TLLLFRKR WQNEKLGLDA GDEYEDENLY EGLNLDDCSM YEDISRGLQG TYQDVGSLNI GDVQLEKPAA AWSHPQFEKG GG SGGGSGG SAWSHPQFEK UniProtKB: B-cell antigen receptor complex-associated protein alpha chain |
-Macromolecule #4: B-cell antigen receptor complex-associated protein beta chain
| Macromolecule | Name: B-cell antigen receptor complex-associated protein beta chain type: protein_or_peptide / ID: 4 / Details: C terminal twin-strep tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 29.163094 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MARLALSPVP SHWMVALLLL LSAEPVPAAR SEDRYRNPKG SACSRIWQSP RFIARKRGFT VKMHCYMNSA SGNVSWLWKQ EMDENPQQL KLEKGRMEES QNESLATLTI QGIRFEDNGI YFCQQKCNNT SEVYQGCGTE LRVMGFSTLA QLKQRNTLKD G IIMIQTLL ...String: MARLALSPVP SHWMVALLLL LSAEPVPAAR SEDRYRNPKG SACSRIWQSP RFIARKRGFT VKMHCYMNSA SGNVSWLWKQ EMDENPQQL KLEKGRMEES QNESLATLTI QGIRFEDNGI YFCQQKCNNT SEVYQGCGTE LRVMGFSTLA QLKQRNTLKD G IIMIQTLL IILFIIVPIF LLLDKDDSKA GMEEDHTYEG LDIDQTATYE DIVTLRTGEV KWSVGEHPGQ EAAAWSHPQF EK GGGSGGG SGGSAWSHPQ FEK UniProtKB: B-cell antigen receptor complex-associated protein beta chain |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 14 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.4000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)