[English] 日本語
 Yorodumi
Yorodumi- EMDB-33352: Cryo-EM structure of occupied ring subunit 4 (OR4) of GroEL compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of occupied ring subunit 4 (OR4) of GroEL complexed with polyalanine model of UGT1A from GroEL-UGT1A double occupied ring complex | ||||||||||||||||||||||||
 Map data Map data | GroEL-UGT1A structure complex EM density map with UGT1A density forming around a single subunit | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | cryogenic electron microscopy / single-particle analysis / molecular motion / structure-function relationship / focus classification / separating heterogeneity / groel / chaperone / unfolded protein | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationGroEL-GroES complex / chaperonin ATPase / virion assembly / : / isomerase activity / ATP-dependent protein folding chaperone / response to radiation / unfolded protein binding / protein folding / response to heat ...GroEL-GroES complex / chaperonin ATPase / virion assembly / : / isomerase activity / ATP-dependent protein folding chaperone / response to radiation / unfolded protein binding / protein folding / response to heat / protein refolding / magnesium ion binding / ATP hydrolysis activity / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | ||||||||||||||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||||||||
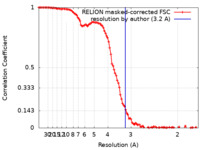
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||||||||||||||
 Authors Authors | Stapleton K / Takagi J / Mizohata E | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Unmasking GroEL: Structure, dynamics, and substrate binding revealed by single-particle cryo-EM Authors: Stapleton KM / Mizobata T / Miyazaki N / Takatsuji T / Kato T / Iwasaki K / Standley DM / Kawamura T / Nakane T / Takagi J / Mizohata E | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33352.map.gz emd_33352.map.gz | 15.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33352-v30.xml emd-33352-v30.xml emd-33352.xml emd-33352.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33352_fsc.xml emd_33352_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_33352.png emd_33352.png | 120.7 KB | ||
| Masks |  emd_33352_msk_1.map emd_33352_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33352.cif.gz emd-33352.cif.gz | 7.4 KB | ||
| Others |  emd_33352_half_map_1.map.gz emd_33352_half_map_1.map.gz emd_33352_half_map_2.map.gz emd_33352_half_map_2.map.gz | 80.8 MB 80.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33352 http://ftp.pdbj.org/pub/emdb/structures/EMD-33352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33352 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33352 | HTTPS FTP |
-Related structure data
| Related structure data |  7xomMC  7xojC  7xokC  7xolC  7xonC  7xooC  7xopC  7xoqC  7xorC  7xosC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33352.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33352.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | GroEL-UGT1A structure complex EM density map with UGT1A density forming around a single subunit | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.87 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33352_msk_1.map emd_33352_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map-2 GroEL-UGT1A structure complex EM density map with UGT1A...
| File | emd_33352_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_map-2__GroEL-UGT1A structure complex EM density map with UGT1A density forming around a single subunit | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map-1 GroEL-UGT1A structure complex EM density map with UGT1A...
| File | emd_33352_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half_map-1__GroEL-UGT1A structure complex EM density map with UGT1A density forming around a single subunit | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GroEL-UGT1A double occupied ring complex
| Entire | Name: GroEL-UGT1A double occupied ring complex |
|---|---|
| Components |
|
-Supramolecule #1: GroEL-UGT1A double occupied ring complex
| Supramolecule | Name: GroEL-UGT1A double occupied ring complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: GroEL Complexed with Unfolded UGT1A reported at 3.2 Ang. global resolution by FSC 0.143 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: OR(4)-GroEL-UGT1A containing modeled (polyalanine) UGT1A
| Supramolecule | Name: OR(4)-GroEL-UGT1A containing modeled (polyalanine) UGT1A type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 / Details: The coordinates of GroEL and UGT1A |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: UDP-glucuronosyltransferase 1A (UGT1A)
| Supramolecule | Name: UDP-glucuronosyltransferase 1A (UGT1A) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 / Details: The model was built |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Chaperonin GroEL
| Macromolecule | Name: Chaperonin GroEL / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO / EC number: chaperonin ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 57.260504 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AAKDVKFGND ARVKMLRGVN VLADAVKVTL GPKGRNVVLD KSFGAPTITK DGVSVAREIE LEDKFENMGA QMVKEVASKA NDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK L IAEAMDKV ...String: AAKDVKFGND ARVKMLRGVN VLADAVKVTL GPKGRNVVLD KSFGAPTITK DGVSVAREIE LEDKFENMGA QMVKEVASKA NDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK L IAEAMDKV GKEGVITVED GTGLQDELDV VEGMQFDRGY LSPYFINKPE TGAVELESPF ILLADKKISN IREMLPVLEA VA KAGKPLL IIAEDVEGEA LATLVVNTMR GIVKVAAVKA PGFGDRRKAM LQDIATLTGG TVISEEIGME LEKATLEDLG QAK RVVINK DTTTIIDGVG EEAAIQGRVA QIRQQIEEAT SDYDREKLQE RVAKLAGGVA VIKVGAATEV EMKEKKARVE DALH ATRAA VEEGVVAGGG VALIRVASKL ADLRGQNEDQ NVGIKVALRA MEAPLRQIVL NCGEEPSVVA NTVKGGDGNY GYNAA TEEY GNMIDMGILD PTKVTRSALQ YAASVAGLMI TTECMVTDLP KNDAADLGAA GGMGGMGGMG GMM UniProtKB: Chaperonin GroEL |
-Macromolecule #2: Polyalanine model of UDP-glucuronosyltransferase 1A (UGT1A)
| Macromolecule | Name: Polyalanine model of UDP-glucuronosyltransferase 1A (UGT1A) type: protein_or_peptide / ID: 2 / Details: Modeled with poly-UNK due to weak density map / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.74137 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 33 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Sample containing GorEL-UGT1A was made fresh and used without undergoing any freeze-thaw cycles to avoid degradation in the solution. The sample was in a buffer solution of 150mM NaCl, 20mM Tris-HCl at pH 7.5 | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: 3 ul of 33 mg/ml GroEL-UGT1A was placed on Holey carbon Quanitifoil copper grids (300 mesh size R1.2/1.3) and blotted for 3 seconds (blot force =1). |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2951 / Average electron dose: 40.0 e/Å2 Details: Using a Titan KRIOS TEM operated at 300 kV, 2,951 movies were collected using the Falcon III DED in Counting mode at a magnification of 75000x corresponding to a pixel size of 0.87 Pixel/A ...Details: Using a Titan KRIOS TEM operated at 300 kV, 2,951 movies were collected using the Falcon III DED in Counting mode at a magnification of 75000x corresponding to a pixel size of 0.87 Pixel/A at the specimen level. All 2,951 movies were imported into the RELION pipeline and prepared for single-particle analysis |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7xom: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)