[English] 日本語
 Yorodumi
Yorodumi- EMDB-33336: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound | |||||||||||||||
 Map data Map data | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | VIRAL PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationMetabolism of Angiotensinogen to Angiotensins / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / peptidyl-dipeptidase activity ...Metabolism of Angiotensinogen to Angiotensins / angiotensin-converting enzyme 2 / positive regulation of L-proline import across plasma membrane / Hydrolases; Acting on peptide bonds (peptidases); Metallocarboxypeptidases / angiotensin-mediated drinking behavior / positive regulation of gap junction assembly / tryptophan transport / regulation of cardiac conduction / maternal process involved in female pregnancy / peptidyl-dipeptidase activity / transporter activator activity / carboxypeptidase activity / angiotensin maturation / metallocarboxypeptidase activity / positive regulation of cardiac muscle contraction / negative regulation of smooth muscle cell proliferation / brush border membrane / negative regulation of ERK1 and ERK2 cascade / virus receptor activity / endopeptidase activity / symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / cilium / symbiont-mediated suppression of host innate immune response / apical plasma membrane / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / cell surface / extracellular space / extracellular region / metal ion binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||||||||
 Authors Authors | Xu Y / Wu C / Liu H / Yin W / Xu HE | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2022 Journal: Cell Res / Year: 2022Title: Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins. Authors: Youwei Xu / Canrong Wu / Xiaodan Cao / Chunyin Gu / Heng Liu / Mengting Jiang / Xiaoxi Wang / Qingning Yuan / Kai Wu / Jia Liu / Deyi Wang / Xianqing He / Xueping Wang / Su-Jun Deng / H Eric Xu / Wanchao Yin /  Abstract: The Omicron BA.2 variant has become a dominant infective strain worldwide. Receptor binding studies show that the Omicron BA.2 spike trimer exhibits 11-fold and 2-fold higher potency in binding to ...The Omicron BA.2 variant has become a dominant infective strain worldwide. Receptor binding studies show that the Omicron BA.2 spike trimer exhibits 11-fold and 2-fold higher potency in binding to human ACE2 than the spike trimer from the wildtype (WT) and Omicron BA.1 strains. The structure of the BA.2 spike trimer complexed with human ACE2 reveals that all three receptor-binding domains (RBDs) in the spike trimer are in open conformation, ready for ACE2 binding, thus providing a basis for the increased infectivity of the BA.2 strain. JMB2002, a therapeutic antibody that was shown to efficiently inhibit Omicron BA.1, also shows potent neutralization activities against Omicron BA.2. In addition, both BA.1 and BA.2 spike trimers are able to bind to mouse ACE2 with high potency. In contrast, the WT spike trimer binds well to cat ACE2 but not to mouse ACE2. The structures of both BA.1 and BA.2 spike trimer bound to mouse ACE2 reveal the basis for their high affinity interactions. Together, these results suggest a possible evolution pathway for Omicron BA.1 and BA.2 variants via a human-cat-mouse-human circle, which could have important implications in establishing an effective strategy for combating SARS-CoV-2 viral infections. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33336.map.gz emd_33336.map.gz | 363.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33336-v30.xml emd-33336-v30.xml emd-33336.xml emd-33336.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33336.png emd_33336.png | 35.8 KB | ||
| Filedesc metadata |  emd-33336.cif.gz emd-33336.cif.gz | 7 KB | ||
| Others |  emd_33336_half_map_1.map.gz emd_33336_half_map_1.map.gz emd_33336_half_map_2.map.gz emd_33336_half_map_2.map.gz | 392 MB 392 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33336 http://ftp.pdbj.org/pub/emdb/structures/EMD-33336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33336 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33336 | HTTPS FTP |
-Validation report
| Summary document |  emd_33336_validation.pdf.gz emd_33336_validation.pdf.gz | 1012 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33336_full_validation.pdf.gz emd_33336_full_validation.pdf.gz | 1011.6 KB | Display | |
| Data in XML |  emd_33336_validation.xml.gz emd_33336_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_33336_validation.cif.gz emd_33336_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33336 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33336 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33336 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33336 | HTTPS FTP |
-Related structure data
| Related structure data |  7xo4MC  7xo5C  7xo6C  7xo7C  7xo8C  7xo9C  7xoaC  7xobC  7xocC  7xodC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33336.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33336.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound | ||||||||||||||||||||||||||||||||||||
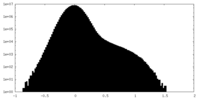
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
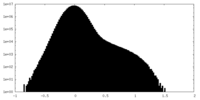
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two...
| File | emd_33336_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
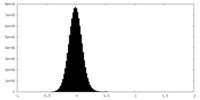
| Density Histograms |
-Half map: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two...
| File | emd_33336_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
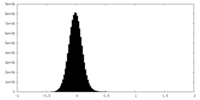
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound
| Entire | Name: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound
| Supramolecule | Name: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with two mouse ACE2 Bound type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer
| Supramolecule | Name: SARS-CoV-2 Omicron BA.1 Variant Spike Trimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: mouse ACE2
| Supramolecule | Name: mouse ACE2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133.911969 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ PFLMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPIIVR EPEDLPQGFS ALEPLVDLPI GINITRFQTL LA LHRSYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFD EVFNATRFAS VYAWNRKRIS NCVADYSVLY NLAPFFTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NKLDSKVSGN YNYLYRLFRK SNLKPFERDI STEIYQAGNK PCNGV AGFN CYFPLRSYSF RPTYGVGHQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLK GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEY VNNSYECDIP IGAGICASYQ TQTKSHGSAS SVASQSIIAY TMSLGAENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLK RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KYFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFKGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTA SALGKLQDVV NHNAQALNTL V KQLSSKFG AISSVLNDIF SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ UniProtKB: Spike glycoprotein |
-Macromolecule #2: Angiotensin-converting enzyme 2
| Macromolecule | Name: Angiotensin-converting enzyme 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO / EC number: angiotensin-converting enzyme 2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.46025 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSSSSWLLLS LVAVTTAQSL TEENAKTFLN NFNQEAEDLS YQSSLASWNY NTNITEENAQ KMSEAAAKWS AFYEEQSKTA QSFSLQEIQ TPIIKRQLQA LQQSGSSALS ADKNKQLNTI LNTMSTIYST GKVCNPKNPQ ECLLLEPGLD EIMATSTDYN S RLWAWEGW ...String: MSSSSWLLLS LVAVTTAQSL TEENAKTFLN NFNQEAEDLS YQSSLASWNY NTNITEENAQ KMSEAAAKWS AFYEEQSKTA QSFSLQEIQ TPIIKRQLQA LQQSGSSALS ADKNKQLNTI LNTMSTIYST GKVCNPKNPQ ECLLLEPGLD EIMATSTDYN S RLWAWEGW RAEVGKQLRP LYEEYVVLKN EMARANNYND YGDYWRGDYE AEGADGYNYN RNQLIEDVER TFAEIKPLYE HL HAYVRRK LMDTYPSYIS PTGCLPAHLL GDMWGRFWTN LYPLTVPFAQ KPNIDVTDAM MNQGWDAERI FQEAEKFFVS VGL PHMTQG FWANSMLTEP ADGRKVVCHP TAWDLGHGDF RIKMCTKVTM DNFLTAHHEM GHIQYDMAYA RQPFLLRNGA NEGF HEAVG EIMSLSAATP KHLKSIGLLP SDFQEDSETE INFLLKQALT IVGTLPFTYM LEKWRWMVFR GEIPKEQWMK KWWEM KREI VGVVEPLPHD ETYCDPASLF HVSNDYSFIR YYTRTIYQFQ FQEALCQAAK YNGSLHKCDI SNSTEAGQKL LKMLSL GNS EPWTKALENV VGARNMDVKP LLNYFQPLFD WLKEQNRNSF VGWNTEWSPY ADQSIKVRIS LKSALGANAY EWTNNEM FL FRSSVAYAMR KYFSIIKNQT VPFLEEDVRV SDLKPRVSFY FFVTSPQNVS DVIPRSEVED AIRMSRGRIN DVFGLNDN S LEFLGIHPTL EPPYQPPVTI WLIIFGVVMA LVVVGIIILI VTGIKGRKKK NETKREENPY DSMDIGKGES NAGFQNSDD AQTSF UniProtKB: Angiotensin-converting enzyme 2 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 39 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.24 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 123465 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7xo4: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)