[English] 日本語
 Yorodumi
Yorodumi- EMDB-33305: E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A filam... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E.coli phosphoribosylpyrophosphate (PRPP) synthetase type A filament bound with ADP, Pi and R5P | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Allosteric enzyme / Kinase / Transferase / Nucleotide biosynthesis / ATP-binding / Magnesium / Manganese / Metal-binding / Nucleotide-binding / BIOSYNTHETIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationribose phosphate diphosphokinase complex / ribonucleoside monophosphate biosynthetic process / ribose-phosphate diphosphokinase / ribose phosphate diphosphokinase activity / 5-phosphoribose 1-diphosphate biosynthetic process / purine nucleotide biosynthetic process / protein hexamerization / phosphate ion binding / ADP binding / kinase activity ...ribose phosphate diphosphokinase complex / ribonucleoside monophosphate biosynthetic process / ribose-phosphate diphosphokinase / ribose phosphate diphosphokinase activity / 5-phosphoribose 1-diphosphate biosynthetic process / purine nucleotide biosynthetic process / protein hexamerization / phosphate ion binding / ADP binding / kinase activity / magnesium ion binding / ATP binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
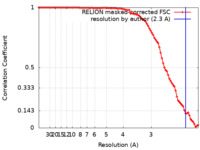
| Method | single particle reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||
 Authors Authors | Hu HH / Lu GM / Chang CC / Liu JL | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: Filamentation modulates allosteric regulation of PRPS. Authors: Huan-Huan Hu / Guang-Ming Lu / Chia-Chun Chang / Yilan Li / Jiale Zhong / Chen-Jun Guo / Xian Zhou / Boqi Yin / Tianyi Zhang / Ji-Long Liu /   Abstract: Phosphoribosyl pyrophosphate (PRPP) is a key intermediate in the biosynthesis of purine and pyrimidine nucleotides, histidine, tryptophan, and cofactors NAD and NADP. Abnormal regulation of PRPP ...Phosphoribosyl pyrophosphate (PRPP) is a key intermediate in the biosynthesis of purine and pyrimidine nucleotides, histidine, tryptophan, and cofactors NAD and NADP. Abnormal regulation of PRPP synthase (PRPS) is associated with human disorders, including Arts syndrome, retinal dystrophy, and gouty arthritis. Recent studies have demonstrated that PRPS can form filamentous cytoophidia in eukaryotes. Here, we show that PRPS forms cytoophidia in prokaryotes both in vitro and in vivo. Moreover, we solve two distinct filament structures of PRPS at near-atomic resolution using Cryo-EM. The formation of the two types of filaments is controlled by the binding of different ligands. One filament type is resistant to allosteric inhibition. The structural comparison reveals conformational changes of a regulatory flexible loop, which may regulate the binding of the allosteric inhibitor and the substrate ATP. A noncanonical allosteric AMP/ADP binding site is identified to stabilize the conformation of the regulatory flexible loop. Our findings not only explore a new mechanism of PRPS regulation with structural basis, but also propose an additional layer of cell metabolism through PRPS filamentation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33305.map.gz emd_33305.map.gz | 39.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33305-v30.xml emd-33305-v30.xml emd-33305.xml emd-33305.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33305_fsc.xml emd_33305_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_33305.png emd_33305.png | 90.8 KB | ||
| Masks |  emd_33305_msk_1.map emd_33305_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33305.cif.gz emd-33305.cif.gz | 6.4 KB | ||
| Others |  emd_33305_half_map_1.map.gz emd_33305_half_map_1.map.gz emd_33305_half_map_2.map.gz emd_33305_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33305 http://ftp.pdbj.org/pub/emdb/structures/EMD-33305 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33305 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33305 | HTTPS FTP |
-Related structure data
| Related structure data |  7xmuMC  7xmvC  7xn3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33305.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33305.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33305_msk_1.map emd_33305_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33305_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_33305_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : E.coli PRPP syhthetase filament structure complex with ADP,Pi, Mg...
| Entire | Name: E.coli PRPP syhthetase filament structure complex with ADP,Pi, Mg2+ and R5P |
|---|---|
| Components |
|
-Supramolecule #1: E.coli PRPP syhthetase filament structure complex with ADP,Pi, Mg...
| Supramolecule | Name: E.coli PRPP syhthetase filament structure complex with ADP,Pi, Mg2+ and R5P type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: a new allosteric for ADP binding |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Ribose-phosphate pyrophosphokinase
| Macromolecule | Name: Ribose-phosphate pyrophosphokinase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: ribose-phosphate diphosphokinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.08409 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPDMKLFAGN ATPELAQRIA NRLYTSLGDA AVGRFSDGEV SVQINENVRG GDIFIIQSTC APTNDNLMEL VVMVDALRRA SAGRITAVI PYFGYARQDR RVRSARVPIT AKVVADFLSS VGVDRVLTVD LHAEQIQGFF DVPVDNVFGS PILLEDMLQL N LDNPIVVS ...String: MPDMKLFAGN ATPELAQRIA NRLYTSLGDA AVGRFSDGEV SVQINENVRG GDIFIIQSTC APTNDNLMEL VVMVDALRRA SAGRITAVI PYFGYARQDR RVRSARVPIT AKVVADFLSS VGVDRVLTVD LHAEQIQGFF DVPVDNVFGS PILLEDMLQL N LDNPIVVS PDIGGVVRAR AIAKLLNDTD MAIIDKRRPR ANVSQVMHII GDVAGRDCVL VDDMIDTGGT LCKAAEALKE RG AKRVFAY ATHPIFSGNA ANNLRNSVID EVVVCDTIPL SDEIKSLPNV RTLTLSGMLA EAIRRISNEE SISAMFEHHH HHH H UniProtKB: Ribose-phosphate pyrophosphokinase |
-Macromolecule #2: 5-O-phosphono-alpha-D-ribofuranose
| Macromolecule | Name: 5-O-phosphono-alpha-D-ribofuranose / type: ligand / ID: 2 / Number of copies: 6 / Formula: HSX |
|---|---|
| Molecular weight | Theoretical: 230.11 Da |
| Chemical component information |  ChemComp-HSX: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 12 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 12 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 822 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: blot for 3.5 seconds with blot force of -1 before plunge-freezing. |
| Details | The purifide monomer or oligomer protein was incubated with ATP and MgCl2 to form this type A filament. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 3474 / Average exposure time: 4.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)