[English] 日本語
 Yorodumi
Yorodumi- EMDB-33240: Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (both in tRNA binding states) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GlyRS2 / Glycine-tRNA ligase / tRNA selection / GlyRS tRNA complex / chloroplasts / LIGASE / LIGASE-RNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationglycyl-tRNA aminoacylation / arginine-tRNA ligase activity / arginyl-tRNA aminoacylation / glycine-tRNA ligase / glycine-tRNA ligase activity / embryo development ending in seed dormancy / regulation of embryonic development / chloroplast / mitochondrion / ATP binding Similarity search - Function | |||||||||
| Biological species |    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Yu Z / Wu Z / Li Y / Lu G / Lin J | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase. Authors: Zhaoli Yu / Zihan Wu / Ye Li / Qiang Hao / Xiaofeng Cao / Gregor M Blaha / Jinzhong Lin / Guoliang Lu /   Abstract: Two types of glycyl-tRNA synthetase (GlyRS) are known, the α2 and the α2β2 GlyRSs. Both types of synthetase employ a class II catalytic domain to aminoacylate tRNAGly. In plastids and some ...Two types of glycyl-tRNA synthetase (GlyRS) are known, the α2 and the α2β2 GlyRSs. Both types of synthetase employ a class II catalytic domain to aminoacylate tRNAGly. In plastids and some bacteria, the α and β subunits are fused and are designated as (αβ)2 GlyRSs. While the tRNA recognition and aminoacylation mechanisms are well understood for α2 GlyRSs, little is known about the mechanisms for α2β2/(αβ)2 GlyRSs. Here we describe structures of the (αβ)2 GlyRS from Oryza sativa chloroplast by itself and in complex with cognate tRNAGly. The set of structures reveals that the U-shaped β half of the synthetase selects the tRNA in a two-step manner. In the first step, the synthetase engages the elbow and the anticodon base C35 of the tRNA. In the second step, the tRNA has rotated ∼9° toward the catalytic centre. The synthetase probes the tRNA for the presence of anticodon base C36 and discriminator base C73. This intricate mechanism enables the tRNA to access the active site of the synthetase from a direction opposite to that of most other class II synthetases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33240.map.gz emd_33240.map.gz | 123.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33240-v30.xml emd-33240-v30.xml emd-33240.xml emd-33240.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33240_fsc.xml emd_33240_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_33240.png emd_33240.png | 68.8 KB | ||
| Masks |  emd_33240_msk_1.map emd_33240_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-33240.cif.gz emd-33240.cif.gz | 6.2 KB | ||
| Others |  emd_33240_half_map_1.map.gz emd_33240_half_map_1.map.gz emd_33240_half_map_2.map.gz emd_33240_half_map_2.map.gz | 171.4 MB 171.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33240 http://ftp.pdbj.org/pub/emdb/structures/EMD-33240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33240 | HTTPS FTP |
-Related structure data
| Related structure data |  7xk1MC  7xjyC  7xjzC  7xk0C  8h1cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33240.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33240.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_33240_msk_1.map emd_33240_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
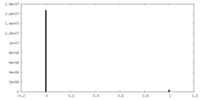
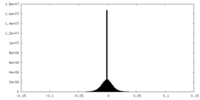
| Density Histograms |
-Half map: #2
| File | emd_33240_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
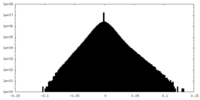
| Density Histograms |
-Half map: #1
| File | emd_33240_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
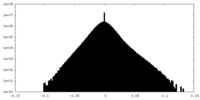
| Density Histograms |
- Sample components
Sample components
-Entire : complex
| Entire | Name: complex |
|---|---|
| Components |
|
-Supramolecule #1: complex
| Supramolecule | Name: complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: glycyl-tRNA synthetase
| Supramolecule | Name: glycyl-tRNA synthetase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: tRNA
| Supramolecule | Name: tRNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glycine--tRNA ligase
| Macromolecule | Name: Glycine--tRNA ligase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: glycine-tRNA ligase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 117.050477 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHGS SLEVLFQGPA VASADGDAPS PVSVSASAAT KGPSSSSVLT FQQAIQRLQD YWASVGCAVM QCSNTEVGAG TMNPLTFLR VLGPEPWNVA YVEPSIRPDD SRYGDNPNRL QRHTQFQVIL KPDPGNSQDL FLHSLSALGI NVREHDIRFV E DNWESPVL ...String: HHHHHHHHGS SLEVLFQGPA VASADGDAPS PVSVSASAAT KGPSSSSVLT FQQAIQRLQD YWASVGCAVM QCSNTEVGAG TMNPLTFLR VLGPEPWNVA YVEPSIRPDD SRYGDNPNRL QRHTQFQVIL KPDPGNSQDL FLHSLSALGI NVREHDIRFV E DNWESPVL GAWGLGWEVW MDGMEITQFT YFQQSGSLPL LPVSVEITYG LERILMSLQG VDHFKNIQYT KGITYGELFL EN EKEMSAY YLEHANVDNI QKHFDDFEEE ARSLLSLWLP IPAYDHVLKA SHAFNILDSR GFVGVTERAR YFGRMRSLAR QCA QLWVKT RENLGYPLGT YQESNLIYPH VSEKPSRKGV VGQPRAFVLE IGTEELPPHD VIEATKQLEK SLIQILEKRR LSHG KVRSY GTPRRLAVVV ENLNMKQMEE EIELRGPPVA KAFDQEGRPT KAAEGFCRKN NVPIDSLYRR TDGKTEYIYA RVKES ARFA DEVLTEDLPT IISGISFPKS MRWNSNIVFS RPIRWIFALH GDLIVPFCFA GISSGNQSCG LRNSSLANFK VEAAEL YLH TLEKAGILID MQERKQRILH DSSILAEGVG GDIIAPDSLV QEVINLVEAP MPIIGRYDVS FLALPKDVLI TVMQKHQ KY FPVTSKTMGN LLPCFITVAN GAIKEEVVRK GNEAVLRARY EDAKFFYKMD TQKKLSEFRD QLSSILFHER LGTMLDKM K RVENTVAEVA LLLGINEKMI PAIKDAAALA MSDLATNIVT EFTSLAGIMA RHYALRDGLS EQIAEALFEI TLPRFSGDV FPKTDPGIVL AVTDRLDSLV GLFGAGCQPS STNDPFGLRR ISYGLVQILV ENKKNFDLTK ALTLVAEEQP ITIDSGVIDE VVQFVTRRL EQLLVDEGIN CEIVRSVLIE RANCPYLASQ TAIEMEAFSR TEDFPKIVEA YSRPTRIIRG KEIGSALEVD A SVFEKDEE RALWSAYLEV ADKIHPGVDI KAFADASLEL LQPLEDFFTN VFVMAEDEKV RNNRLALLTK VASLPKGIAD LS VLPGF UniProtKB: glycine--tRNA ligase |
-Macromolecule #2: tRNA(gly)
| Macromolecule | Name: tRNA(gly) / type: rna / ID: 2 / Number of copies: 2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.906104 KDa |
| Sequence | String: (GTP)CGAGCGUAG UUCAAUGGUA AAACAUCUCC UUGCCAAGGA GAAGAUACGG GUUCGAUUCC CGCCGCUCGC CCCA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)