[English] 日本語
 Yorodumi
Yorodumi- EMDB-33216: Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human TRPV3_G573S in complex with Trpvicin in C2 symmetry | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcalcium channel activity / lysosome / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.93 Å | |||||||||
 Authors Authors | Fan J / Yue Z / Jiang D / Lei X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Structural basis of TRPV3 inhibition by an antagonist. Authors: Junping Fan / Linghan Hu / Zongwei Yue / Daohong Liao / Fusheng Guo / Han Ke / Daohua Jiang / Yong Yang / Xiaoguang Lei /  Abstract: The TRPV3 channel plays vital roles in skin physiology. Dysfunction of TRPV3 causes skin diseases, including Olmsted syndrome. However, the lack of potent and selective inhibitors impedes the ...The TRPV3 channel plays vital roles in skin physiology. Dysfunction of TRPV3 causes skin diseases, including Olmsted syndrome. However, the lack of potent and selective inhibitors impedes the validation of TRPV3 as a therapeutic target. In this study, we identified Trpvicin as a potent and subtype-selective inhibitor of TRPV3. Trpvicin exhibits pharmacological potential in the inhibition of itch and hair loss in mouse models. Cryogenic electron microscopy structures of TRPV3 and the pathogenic G573S mutant complexed with Trpvicin reveal detailed ligand-binding sites, suggesting that Trpvicin inhibits the TRPV3 channel by stabilizing it in a closed state. Our G573S mutant structures demonstrate that the mutation causes a dilated pore, generating constitutive opening activity. Trpvicin accesses additional binding sites inside the central cavity of the G573S mutant to remodel the channel symmetry and block the channel. Together, our results provide mechanistic insights into the inhibition of TRPV3 by Trpvicin and support TRPV3-related drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33216.map.gz emd_33216.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33216-v30.xml emd-33216-v30.xml emd-33216.xml emd-33216.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33216.png emd_33216.png | 74.6 KB | ||
| Filedesc metadata |  emd-33216.cif.gz emd-33216.cif.gz | 6.8 KB | ||
| Others |  emd_33216_half_map_1.map.gz emd_33216_half_map_1.map.gz emd_33216_half_map_2.map.gz emd_33216_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33216 http://ftp.pdbj.org/pub/emdb/structures/EMD-33216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33216 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33216 | HTTPS FTP |
-Related structure data
| Related structure data |  7xj1MC  7xj0C  7xj2C  7xj3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33216.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33216.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33216_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
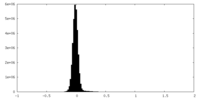
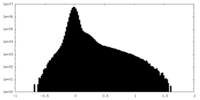
| Density Histograms |
-Half map: #1
| File | emd_33216_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
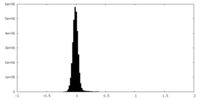
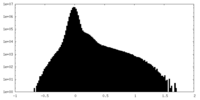
| Density Histograms |
- Sample components
Sample components
-Entire : TRPV3 in complex with an antagonist
| Entire | Name: TRPV3 in complex with an antagonist |
|---|---|
| Components |
|
-Supramolecule #1: TRPV3 in complex with an antagonist
| Supramolecule | Name: TRPV3 in complex with an antagonist / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: Fusion protein of Transient receptor potential cation channel sub...
| Macromolecule | Name: Fusion protein of Transient receptor potential cation channel subfamily V member 3 and 3C-GFP type: protein_or_peptide / ID: 1 Details: The domain (792-799) is PreScission Site. The rest domain (800-1033) is corresponding to this sfGFP (2-235 amino acids).,The domain (792-799) is PreScission Site. The rest domain (800-1033) ...Details: The domain (792-799) is PreScission Site. The rest domain (800-1033) is corresponding to this sfGFP (2-235 amino acids).,The domain (792-799) is PreScission Site. The rest domain (800-1033) is corresponding to this sfGFP (2-235 amino acids). Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 120.865406 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKAHPKEMVP LMGKRVAAPS GNPAILPEKR PAEITPTKKS AHFFLEIEGF EPNPTVAKTS PPVFSKPMDS NIRQCISGNC DDMDSPQSP QDDVTETPSN PNSPSAQLAK EEQRRKKRRL KKRIFAAVSE GCVEELVELL VELQELCRRR HDEDVPDFLM H KLTASDTG ...String: MKAHPKEMVP LMGKRVAAPS GNPAILPEKR PAEITPTKKS AHFFLEIEGF EPNPTVAKTS PPVFSKPMDS NIRQCISGNC DDMDSPQSP QDDVTETPSN PNSPSAQLAK EEQRRKKRRL KKRIFAAVSE GCVEELVELL VELQELCRRR HDEDVPDFLM H KLTASDTG KTCLMKALLN INPNTKEIVR ILLAFAEEND ILGRFINAEY TEEAYEGQTA LNIAIERRQG DIAALLIAAG AD VNAHAKG AFFNPKYQHE GFYFGETPLA LAACTNQPEI VQLLMEHEQT DITSRDSRGN NILHALVTVA EDFKTQNDFV KRM YDMILL RSGNWELETT RNNDGLTPLQ LAAKMGKAEI LKYILSREIK EKRLRSLSRK FTDWAYGPVS SSLYDLTNVD TTTD NSVLE ITVYNTNIDN RHEMLTLEPL HTLLHMKWKK FAKHMFFLSF CFYFFYNITL TLVSYYRPRE EEAIPHPLAL THKMG WLQL LGRMFVLIWA MCISVKEGIA IFLLRPSDLQ SILSDAWFHF VFFIQAVLVI LSVFLYLFAY KEYLACLVLA MALGWA NML YYTRGFQSMS MYSVMIQKVI LHDVLKFLFV YIVFLLGFGV ALASLIEKCP KDNKDCSSYG SFSDAVLELF KLTIGLG DL NIQQNSKYPI LFLFLLITYV ILTFVLLLNM LIALMGETVE NVSKESERIW RLQRARTILE FEKMLPEWLR SRFRMGEL C KVAEDDFRLC LRINEVKWTE WKTHVSFLNE DPGPVRRTAD FNKIQDSSRN NSKTTLNAFE EVEEFPETSV LEVLFQGPS KGEELFTGVV PILVELDGDV NGHKFSVRGE GEGDATNGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKRHD FFKSAMPEG YVQERTISFK DDGTYKTRAE VKFEGDTLVN RIELKGIDFK EDGNILGHKL EYNFNSHNVY ITADKQKNGI K ANFKIRHN VEDGSVQLAD HYQQNTPIGD GPVLLPDNHY LSTQSVLSKD PNEKRDHMVL LEFVTAAGIT HGMDEWSHPQ FE KGGGSGG GSGGSAWSHP QFEK UniProtKB: Transient receptor potential cation channel subfamily V member 3 |
-Macromolecule #2: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyl...
| Macromolecule | Name: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate type: ligand / ID: 2 / Number of copies: 10 / Formula: 6OU |
|---|---|
| Molecular weight | Theoretical: 717.996 Da |
| Chemical component information |  ChemComp-6OU: |
-Macromolecule #3: N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3...
| Macromolecule | Name: N-[5-[2-(2-cyanopropan-2-yl)pyridin-4-yl]-4-(trifluoromethyl)-1,3-thiazol-2-yl]-4,6-dimethoxy-pyrimidine-5-carboxamide type: ligand / ID: 3 / Number of copies: 10 / Formula: EQK |
|---|---|
| Molecular weight | Theoretical: 478.448 Da |
| Chemical component information | 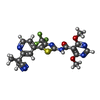 ChemComp-EQK: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


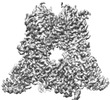





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)