[English] 日本語
 Yorodumi
Yorodumi- EMDB-33199: Cryo-EM structure of the tubular assembly of WGA protein without ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the tubular assembly of WGA protein without symmetry imposition | |||||||||
 Map data Map data | cryo-EM map of wheat germ agglutinin protein tube witout symmetry imposition | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | protein tube / self-assembly / carbohydrate interaction / PLANT PROTEIN | |||||||||
| Biological species |  | |||||||||
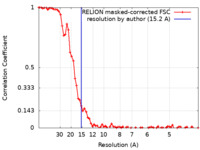
| Method | helical reconstruction / cryo EM / Resolution: 15.2 Å | |||||||||
 Authors Authors | Zhang L / Chen SY / Liu RY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: ACS Nano / Year: 2023 Journal: ACS Nano / Year: 2023Title: Evolution of Protein Assemblies Driven by the Switching of Interplay Mode. Authors: Rongying Liu / Long Li / Shuyu Chen / Zhiwei Yang / Zdravko Kochovski / Shilin Mei / Yan Lu / Lei Zhang / Guosong Chen /   Abstract: A protein assembly with the ability to switch interplay modes of multiple driving forces has been achieved. Although biomolecular systems driven by multiple driving forces have been exploited, work ...A protein assembly with the ability to switch interplay modes of multiple driving forces has been achieved. Although biomolecular systems driven by multiple driving forces have been exploited, work on such a protein assembly capable of switching the interplay modes at nanoscale has been rarely reported so far as a result of their great fabrication challenge. In this work, two sets of driving forces such as ligand-ligand interaction and protein-protein interaction were leveraged to antagonistically underpin the multilayered stackings and trigger the hollow evolution to afford the well-defined hollow rectangular frame of proteins. While these protein frames further collapsed into aggregates, the ligand-ligand interactions were weakened, and the interplay of two sets of driving forces thereby tended to switch into synergistic mode, converting the protein packing mode from porously loose packing to axially dense packing and thus giving rise to a morphological evolution toward a nanosized protein tube. This strategy not only provides a nanoscale understanding on the mechanism underlying the switch of interplay modes in the context of biomacromolecules but also may provide access for diverse sophisticated biomacromolecular nanostructures that are historically inaccessible for conventional self-assembly strategies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33199.map.gz emd_33199.map.gz | 27.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33199-v30.xml emd-33199-v30.xml emd-33199.xml emd-33199.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33199_fsc.xml emd_33199_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_33199.png emd_33199.png | 73.2 KB | ||
| Others |  emd_33199_half_map_1.map.gz emd_33199_half_map_1.map.gz emd_33199_half_map_2.map.gz emd_33199_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33199 http://ftp.pdbj.org/pub/emdb/structures/EMD-33199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33199 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33199 | HTTPS FTP |
-Validation report
| Summary document |  emd_33199_validation.pdf.gz emd_33199_validation.pdf.gz | 812.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33199_full_validation.pdf.gz emd_33199_full_validation.pdf.gz | 812.4 KB | Display | |
| Data in XML |  emd_33199_validation.xml.gz emd_33199_validation.xml.gz | 12.8 KB | Display | |
| Data in CIF |  emd_33199_validation.cif.gz emd_33199_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33199 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33199 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33199 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33199 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33199.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33199.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of wheat germ agglutinin protein tube witout symmetry imposition | ||||||||||||||||||||
| Voxel size | X=Y=Z: 2.051 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EM half map of wheat germ agglutinin protein tube
| File | emd_33199_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map of wheat germ agglutinin protein tube | ||||||||||||
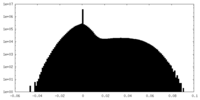
| Projections & Slices |
| ||||||||||||
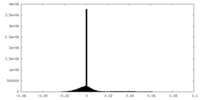
| Density Histograms |
-Half map: EM half map of wheat germ agglutinin protein tube
| File | emd_33199_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map of wheat germ agglutinin protein tube | ||||||||||||
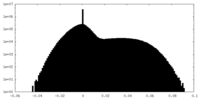
| Projections & Slices |
| ||||||||||||
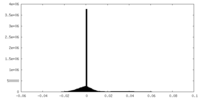
| Density Histograms |
- Sample components
Sample components
-Entire : Helical complex of wheat germ agglutinin protein with R-SL ligand
| Entire | Name: Helical complex of wheat germ agglutinin protein with R-SL ligand |
|---|---|
| Components |
|
-Supramolecule #1: Helical complex of wheat germ agglutinin protein with R-SL ligand
| Supramolecule | Name: Helical complex of wheat germ agglutinin protein with R-SL ligand type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI CETA (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z
Z Y
Y X
X