[English] 日本語
 Yorodumi
Yorodumi- EMDB-33190: Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VIRUS LIKE PARTICLE | |||||||||
| Function / homology | virion component / Viral coat protein subunit / Capsid protein alpha Function and homology information Function and homology information | |||||||||
| Biological species |  Lake Sinai virus 2 Lake Sinai virus 2 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | |||||||||
 Authors Authors | Chen NC / Wang CH / Chen CJ / Yoshimura M / Guan HH / Chuankhayan P / Lin CC | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions Authors: Chen NC / Wang CH / Yoshimura M / Yeh YQ / Guan HH / Chuankhayan P / Lin CC / Lin PJ / Huang YC / Wakatsuki S / Ho MC / Chen CJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33190.map.gz emd_33190.map.gz | 483.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33190-v30.xml emd-33190-v30.xml emd-33190.xml emd-33190.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33190.png emd_33190.png | 143.5 KB | ||
| Filedesc metadata |  emd-33190.cif.gz emd-33190.cif.gz | 5.7 KB | ||
| Others |  emd_33190_half_map_1.map.gz emd_33190_half_map_1.map.gz emd_33190_half_map_2.map.gz emd_33190_half_map_2.map.gz | 474 MB 474 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33190 http://ftp.pdbj.org/pub/emdb/structures/EMD-33190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33190 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33190 | HTTPS FTP |
-Validation report
| Summary document |  emd_33190_validation.pdf.gz emd_33190_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33190_full_validation.pdf.gz emd_33190_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_33190_validation.xml.gz emd_33190_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_33190_validation.cif.gz emd_33190_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33190 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33190 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33190 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33190 | HTTPS FTP |
-Related structure data
| Related structure data |  7xgzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33190.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33190.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0537 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33190_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
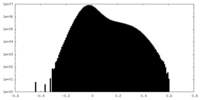
| Projections & Slices |
| ||||||||||||
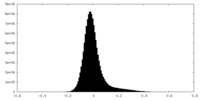
| Density Histograms |
-Half map: #2
| File | emd_33190_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
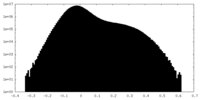
| Projections & Slices |
| ||||||||||||
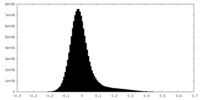
| Density Histograms |
- Sample components
Sample components
-Entire : Lake Sinai virus 2
| Entire | Name:  Lake Sinai virus 2 Lake Sinai virus 2 |
|---|---|
| Components |
|
-Supramolecule #1: Lake Sinai virus 2
| Supramolecule | Name: Lake Sinai virus 2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1041831 / Sci species name: Lake Sinai virus 2 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|
-Macromolecule #1: Capsid protein alpha
| Macromolecule | Name: Capsid protein alpha / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lake Sinai virus 2 Lake Sinai virus 2 |
| Molecular weight | Theoretical: 57.344309 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNPPTTTTTT TRTIRAPKVQ LTPNSATRRR RNRRRRRPAA AIAGPLSIQP SSINRRMVSR VTRRSAVVTA AGLAWLRQYL NPMGPDTTS VTGYPDGSAV TTCIADYSNT FNVSFPPREA LYCTGSSSSE KPTLVDADNY AKIDKWSNYD ITLCVLALPM L RNVVMLRL ...String: MNPPTTTTTT TRTIRAPKVQ LTPNSATRRR RNRRRRRPAA AIAGPLSIQP SSINRRMVSR VTRRSAVVTA AGLAWLRQYL NPMGPDTTS VTGYPDGSAV TTCIADYSNT FNVSFPPREA LYCTGSSSSE KPTLVDADNY AKIDKWSNYD ITLCVLALPM L RNVVMLRL YPHTPTAFAL TEQTPNFPQR FPNWSVYSAD GTRFNNGDEP GYLQSYVYLP NVDKHLSAAR GYRLLSRGIT GI FSAPALE TQGFVTACQY LAEGSIQSQS IKSDAVRSVT VNSDGTVKNV ESSSQTVSSM PRYVFPLDGD NCAPSSLTET YHQ AYQSKA TDGFYMPVLS SSRDNPFHPP QPRAIAVYGS FLARGCLDPV SEAHEADGPT HDIYRLNVAD DVAPLFNTGV VWFE GISPK FSLKLKTRTV LQYIPTSGSV LANFTRHEPT YDQIALDAAD RLRNLMPHAY PAAYNDWGWL GDLLDSAISM LPGVG TVYN IAKPLIKPAW NWLGNKVSDF FGNPVARDGD IFFDAK UniProtKB: Capsid protein alpha |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 46.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.72 µm / Nominal defocus min: 0.25 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


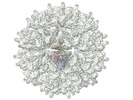
 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)