+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full length human CLC-2 channel in apo state | |||||||||
 Map data Map data | full lenth human CLC-2 channel in apo state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | homo-dimer / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of aldosterone biosynthetic process / cell differentiation involved in salivary gland development / regulation of membrane depolarization during action potential / volume-sensitive chloride channel activity / astrocyte end-foot / stabilization of membrane potential / acinar cell differentiation / cellular hypotonic response / regulation of resting membrane potential / voltage-gated chloride channel activity ...regulation of aldosterone biosynthetic process / cell differentiation involved in salivary gland development / regulation of membrane depolarization during action potential / volume-sensitive chloride channel activity / astrocyte end-foot / stabilization of membrane potential / acinar cell differentiation / cellular hypotonic response / regulation of resting membrane potential / voltage-gated chloride channel activity / axon initial segment / chloride channel regulator activity / dendritic spine membrane / chloride transport / phagocytosis, engulfment / positive regulation of oligodendrocyte differentiation / chloride channel complex / lung development / Stimuli-sensing channels / myelin sheath / retina development in camera-type eye / perikaryon / basolateral plasma membrane / postsynaptic membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
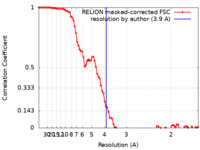
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Wang L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42 Authors: Ma T / Wang L / Chai A / Liu C / Cui W / Yuan S / Wing Ngor Au S / Sun L / Zhang X / Zhang Z / Lu J / Gao Y / Wang P / Li Z / Liang Y / Vogel H / Wang YT / Wang D / Yan K / Zhang H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33169.map.gz emd_33169.map.gz | 6.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33169-v30.xml emd-33169-v30.xml emd-33169.xml emd-33169.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33169_fsc.xml emd_33169_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_33169.png emd_33169.png | 50.7 KB | ||
| Filedesc metadata |  emd-33169.cif.gz emd-33169.cif.gz | 6 KB | ||
| Others |  emd_33169_half_map_1.map.gz emd_33169_half_map_1.map.gz emd_33169_half_map_2.map.gz emd_33169_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33169 http://ftp.pdbj.org/pub/emdb/structures/EMD-33169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33169 | HTTPS FTP |
-Validation report
| Summary document |  emd_33169_validation.pdf.gz emd_33169_validation.pdf.gz | 793.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33169_full_validation.pdf.gz emd_33169_full_validation.pdf.gz | 793.4 KB | Display | |
| Data in XML |  emd_33169_validation.xml.gz emd_33169_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_33169_validation.cif.gz emd_33169_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33169 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33169 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33169 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33169 | HTTPS FTP |
-Related structure data
| Related structure data |  7xf5MC  7xjaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33169.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33169.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full lenth human CLC-2 channel in apo state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.829 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_33169_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_33169_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Full length human CLC-2 homo dimer
| Entire | Name: Full length human CLC-2 homo dimer |
|---|---|
| Components |
|
-Supramolecule #1: Full length human CLC-2 homo dimer
| Supramolecule | Name: Full length human CLC-2 homo dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 180 kDa/nm |
-Macromolecule #1: Chloride channel protein 2
| Macromolecule | Name: Chloride channel protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 98.642352 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAAAAEEGM EPRALQYEQT LMYGRYTQDL GAFAKEEAAR IRLGGPEPWK GPPSSRAAPE LLEYGRSRCA RCRVCSVRCH KFLVSRVGE DWIFLVLLGL LMALVSWVMD YAIAACLQAQ QWMSRGLNTS ILLQYLAWVT YPVVLITFSA GFTQILAPQA V GSGIPEMK ...String: MAAAAAEEGM EPRALQYEQT LMYGRYTQDL GAFAKEEAAR IRLGGPEPWK GPPSSRAAPE LLEYGRSRCA RCRVCSVRCH KFLVSRVGE DWIFLVLLGL LMALVSWVMD YAIAACLQAQ QWMSRGLNTS ILLQYLAWVT YPVVLITFSA GFTQILAPQA V GSGIPEMK TILRGVVLKE YLTLKTFIAK VIGLTCALGS GMPLGKEGPF VHIASMCAAL LSKFLSLFGG IYENESRNTE ML AAACAVG VGCCFAAPIG GVLFSIEVTS TFFAVRNYWR GFFAATFSAF IFRVLAVWNR DEETITALFK TRFRLDFPFD LQE LPAFAV IGIASGFGGA LFVYLNRKIV QVMRKQKTIN RFLMRKRLLF PALVTLLIST LTFPPGFGQF MAGQLSQKET LVTL FDNRT WVRQGLVEEL EPPSTSQAWN PPRANVFLTL VIFILMKFWM SALATTIPVP CGAFMPVFVI GAAFGRLVGE SMAAW FPDG IHTDSSTYRI VPGGYAVVGA AALAGAVTHT VSTAVIVFEL TGQIAHILPV MIAVILANAV AQSLQPSLYD SIIRIK KLP YLPELGWGRH QQYRVRVEDI MVRDVPHVAL SCTFRDLRLA LHRTKGRMLA LVESPESMIL LGSIERSQVV ALLGAQL SP ARRRQHMQER RATQTSPLSD QEGPPTPEAS VCFQVNTEDS AFPAARGETH KPLKPALKRG PSVTRNLGES PTGSAESA G IALRSLFCGS PPPEAASEKL ESCEKRKLKR VRISLASDAD LEGEMSPEEI LEWEEQQLDE PVNFSDCKID PAPFQLVER TSLHKTHTIF SLLGVDHAYV TSIGRLIGIV TLKELRKAIE GSVTAQGVKV RPPLASFRDS ATSSSDTETT EVHALWGPHS RHGLPREGS PSDSDDKCQ UniProtKB: Chloride channel protein 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 300 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7xf5: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)