+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tail structure of bacteriophage SSV19 | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Complex / VIRAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcarbohydrate metabolic process / structural molecule activity / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |   Sulfolobus spindle-shaped virus Sulfolobus spindle-shaped virus | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||
 Authors Authors | Liu HR / Chen WY | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structural insights into a spindle-shaped archaeal virus with a sevenfold symmetrical tail. Authors: Zhen Han / Wanjuan Yuan / Hao Xiao / Li Wang / Junxia Zhang / Yuning Peng / Lingpeng Cheng / Hongrong Liu / Li Huang /  Abstract: Archaeal viruses with a spindle-shaped virion are abundant and widespread in extremely diverse environments. However, efforts to obtain the high-resolution structure of a spindle-shaped virus have ...Archaeal viruses with a spindle-shaped virion are abundant and widespread in extremely diverse environments. However, efforts to obtain the high-resolution structure of a spindle-shaped virus have been unsuccessful. Here, we present the structure of SSV19, a spindle-shaped virus infecting the hyperthermophilic archaeon sp. E11-6. Our near-atomic structure reveals an unusual sevenfold symmetrical virus tail consisting of the tailspike, nozzle, and adaptor proteins. The spindle-shaped capsid shell is formed by seven left-handed helical strands, constructed of the hydrophobic major capsid protein, emanating from the highly glycosylated tail assembly. Sliding between adjacent strands is responsible for the variation of a virion in size. Ultrathin sections of the SSV19-infected cells show that SSV19 virions adsorb to the host cell membrane through the tail after penetrating the S-layer. The tailspike harbors a putative endo-mannanase domain, which shares structural similarity to a endo-mannanase. Molecules of glycerol dibiphytanyl glycerol tetraether lipid were observed in hydrophobic clefts between the tail and the capsid shell. The nozzle protein resembles the stem and clip domains of the portals of herpesviruses and bacteriophages, implying an evolutionary relationship among the archaeal, bacterial, and eukaryotic viruses. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33148.map.gz emd_33148.map.gz | 227.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33148-v30.xml emd-33148-v30.xml emd-33148.xml emd-33148.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33148.png emd_33148.png | 65.2 KB | ||
| Filedesc metadata |  emd-33148.cif.gz emd-33148.cif.gz | 6.1 KB | ||
| Others |  emd_33148_half_map_1.map.gz emd_33148_half_map_1.map.gz emd_33148_half_map_2.map.gz emd_33148_half_map_2.map.gz | 92.3 MB 92.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33148 http://ftp.pdbj.org/pub/emdb/structures/EMD-33148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33148 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33148 | HTTPS FTP |
-Validation report
| Summary document |  emd_33148_validation.pdf.gz emd_33148_validation.pdf.gz | 866.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33148_full_validation.pdf.gz emd_33148_full_validation.pdf.gz | 866.3 KB | Display | |
| Data in XML |  emd_33148_validation.xml.gz emd_33148_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_33148_validation.cif.gz emd_33148_validation.cif.gz | 19.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33148 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33148 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33148 | HTTPS FTP |
-Related structure data
| Related structure data |  7xdiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33148.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33148.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.27 Å | ||||||||||||||||||||||||||||||||||||
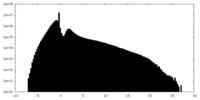
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33148_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
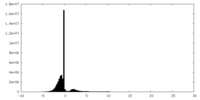
| Density Histograms |
-Half map: #1
| File | emd_33148_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
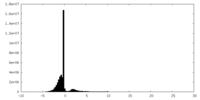
| Density Histograms |
- Sample components
Sample components
-Entire : Sulfolobus spindle-shaped virus
| Entire | Name:   Sulfolobus spindle-shaped virus Sulfolobus spindle-shaped virus |
|---|---|
| Components |
|
-Supramolecule #1: Sulfolobus spindle-shaped virus
| Supramolecule | Name: Sulfolobus spindle-shaped virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 2491899 / Sci species name: Sulfolobus spindle-shaped virus / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: VP1
| Macromolecule | Name: VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Sulfolobus spindle-shaped virus Sulfolobus spindle-shaped virus |
| Molecular weight | Theoretical: 9.282206 KDa |
| Sequence | String: MRWQIRKLRR AEGFNVGLLI GLFIFILVGI VLLPVITSEV SSLTSGTSPA VTGTNATLLN LVPLFYILIL VIVPAVLAYK MYRE UniProtKB: VP1 |
-Macromolecule #2: C131
| Macromolecule | Name: C131 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Sulfolobus spindle-shaped virus Sulfolobus spindle-shaped virus |
| Molecular weight | Theoretical: 14.753507 KDa |
| Sequence | String: MGTKIIINVI FFDIILALLM MSFASIQPPS IANPPTVAQA QAQANITWNL TVGSINWEWL WPVFYFVDWL IWIVTTIFAV VAFIFNVFT TSLSLLASVP IVGPFLLMFA VIINFVLIWE VVKLIRGYDN PG UniProtKB: Uncharacterized protein |
-Macromolecule #3: B210
| Macromolecule | Name: B210 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Sulfolobus spindle-shaped virus Sulfolobus spindle-shaped virus |
| Molecular weight | Theoretical: 23.195996 KDa |
| Sequence | String: MKWPLLLFTV LLIIGFTLIA RAGTISLLST PPVNPPAYSY FYIEFQFLPT NNTPQPYAIF VGPNPNNLTE VAEGYTLSNG TGYARVPVI NAQTEYVDIV WVNQNYTMFE IFPQIQNATT TVTLSANNNQ GFSFSLPTWV SWVIGAVLML IFMGVGWKFM G PAGLAIFG ...String: MKWPLLLFTV LLIIGFTLIA RAGTISLLST PPVNPPAYSY FYIEFQFLPT NNTPQPYAIF VGPNPNNLTE VAEGYTLSNG TGYARVPVI NAQTEYVDIV WVNQNYTMFE IFPQIQNATT TVTLSANNNQ GFSFSLPTWV SWVIGAVLML IFMGVGWKFM G PAGLAIFG IFGLFIAMFF GLLPSYLMYV ILFIVAIVGA RILTKQLGGG EE UniProtKB: Uncharacterized protein |
-Macromolecule #4: VP4
| Macromolecule | Name: VP4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Sulfolobus spindle-shaped virus Sulfolobus spindle-shaped virus |
| Molecular weight | Theoretical: 137.071953 KDa |
| Sequence | String: MKRVFLLYII GILLTLFLPL IQTQSAVSLP PLYVEDAVNA EIQQLWSKSP TGVYAFHEAP SVNNSFWPDD NAKFLESIAP WWQSYSSYV NSTLQFLQQS DVNGLFIKRF EYPLNPLQSI TIGNLSGYTN GFYDIVGNPL LNSMRIATYY NPTLAVTYLF G NVVQYPNG ...String: MKRVFLLYII GILLTLFLPL IQTQSAVSLP PLYVEDAVNA EIQQLWSKSP TGVYAFHEAP SVNNSFWPDD NAKFLESIAP WWQSYSSYV NSTLQFLQQS DVNGLFIKRF EYPLNPLQSI TIGNLSGYTN GFYDIVGNPL LNSMRIATYY NPTLAVTYLF G NVVQYPNG ILVNIEQGLE NPITDGGFGG TGGQNPPWES LNSSSLVNDS IVSIVNNAKT YLNLTGPTFF GTPSEELQYN FP IVNVLPH YLAFQNVNGI LGQYNYQGKF IPFNVTLVLQ SSSINRIYLE FIWENSTSGT YVLTDIPVYF TANGQWQQVV VTV PASAWP KYWNLGALSA VPLLIGIGLD LPGSSPSQTG PTGVYVGDIA TNYPTTFGPQ FNVTNKGSYV VFNESWKSDS LGAT FWIAY VLGQGNAIEV LASAPVNQSW IYVGYNGLAT IGTGYTILET PSGILKNYQN SGNISWTYLG PNFGKWMLLS TNYAP NWIG DFQMLFIFPM AGTSNPYMDT LNNAVYMGDP TEVRNTLYFG NYTTLPGYFQ WVQIAYQNDG NTSGVFGFFL IPSVDY LVN PSVIVNDMFP SSLTAYSPSS IPNYWWEAVW GENYYEGEII YALALLGKYG NSQALQMAQQ AWLSYYNQLK AYNGATY TS SLARFIMATI LLYNITGNTQ YSNAYTQLAN WLLQYQNQSK YAYVYIPMWY HKDVDVPSVN GFATYGYIIN RTAQMDVG T VISGTSIGLN FFEDIPLNTS YGIYLLTNGT GKLPFTYQNV LNVSGTFITY LYMNGGGTAT TANITITVQI AYNGNVLQT IGTAAVDNVP IQPGGISGSP PFYPVKIVVP VLTTVNAPPG STLIIGWNIK APQTVYVLID STNGPSNVTI PLSWPNPFYG LFTIPKIYN PNPGVHNYPQ PYFLDISAMA GQAMMALYAV TKNITYLLDA QLVMNAIHYG PVPMPTYGIL GVPNPPVEPR L WVYANYST VDADYYTYKS ELVSEFGDAI GNNTLASLAI SRVWQRTSYT YPTSYIYYVA RYGSGLQMNS ETQPWGDVAT QF YVNTWSP SNLDLFWASL PNNNYITNQT WNGTALFIHL YAYQQSQVQL IFLTTTVNFN VLVNGNYTNY EANHQIMQIA PTL EPGPNT IIIIPNPKNQ VSQNTNISTT TTTSPLSNAI SGLGITLTQN ELMLLGFVIY FVIIMVTYGV SRNKTITVLS SIVA VAIVY ALALWPTYMA FILGAVGFFM LFYSISRREE E UniProtKB: VP4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 49361 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)