[English] 日本語
 Yorodumi
Yorodumi- EMDB-33145: Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structures of human mitochondrial NAD(P)+-dependent malic enzyme in apo form | |||||||||
 Map data Map data | ME2-APO Full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Human mitochondrial NAD(P)+-dependent malic enzyme (ME2) / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmalate dehydrogenase (oxaloacetate-decarboxylating) / regulation of NADP metabolic process / malate dehydrogenase (decarboxylating) (NAD+) activity / malic enzyme activity / malate dehydrogenase (decarboxylating) (NADP+) activity / oxaloacetate decarboxylase activity / malate metabolic process / pyruvate metabolic process / Pyruvate metabolism / Mitochondrial protein degradation ...malate dehydrogenase (oxaloacetate-decarboxylating) / regulation of NADP metabolic process / malate dehydrogenase (decarboxylating) (NAD+) activity / malic enzyme activity / malate dehydrogenase (decarboxylating) (NADP+) activity / oxaloacetate decarboxylase activity / malate metabolic process / pyruvate metabolic process / Pyruvate metabolism / Mitochondrial protein degradation / NAD binding / electron transfer activity / mitochondrial matrix / intracellular membrane-bounded organelle / mitochondrion / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.72 Å | |||||||||
 Authors Authors | Wang CH / Hsieh JT / Ho MC / Hung HC | |||||||||
| Funding support |  Taiwan, 2 items Taiwan, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Suppression of the human malic enzyme 2 modifies energy metabolism and inhibits cellular respiration Authors: Hsieh JY / Chen KC / Wang CH / Liu GY / Ye JA / Chou YT / Lin YC / Lyu CJ / Chang RY / Liu YL / Li YH / Lee MR / Ho MC / Hung HC | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33145.map.gz emd_33145.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33145-v30.xml emd-33145-v30.xml emd-33145.xml emd-33145.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33145_fsc.xml emd_33145_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_33145.png emd_33145.png | 209 KB | ||
| Filedesc metadata |  emd-33145.cif.gz emd-33145.cif.gz | 5.5 KB | ||
| Others |  emd_33145_half_map_1.map.gz emd_33145_half_map_1.map.gz emd_33145_half_map_2.map.gz emd_33145_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33145 http://ftp.pdbj.org/pub/emdb/structures/EMD-33145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33145 | HTTPS FTP |
-Related structure data
| Related structure data |  7xdeMC  7xdfC  7xdgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ME2-APO Full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: ME2-APO Half map 1
| File | emd_33145_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ME2-APO Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
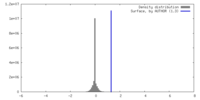
| Density Histograms |
-Half map: ME2-APO Half map 2
| File | emd_33145_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ME2-APO Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
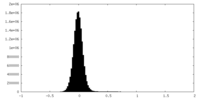
| Density Histograms |
- Sample components
Sample components
-Entire : human mitochondrial NAD(P)+-dependent malic enzyme in apo form
| Entire | Name: human mitochondrial NAD(P)+-dependent malic enzyme in apo form |
|---|---|
| Components |
|
-Supramolecule #1: human mitochondrial NAD(P)+-dependent malic enzyme in apo form
| Supramolecule | Name: human mitochondrial NAD(P)+-dependent malic enzyme in apo form type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: NAD-dependent malic enzyme, mitochondrial
| Macromolecule | Name: NAD-dependent malic enzyme, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO EC number: malate dehydrogenase (oxaloacetate-decarboxylating) |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.521434 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLSRLRVVST TCTLACRHLH IKEKGKPLML NPRTNKGMAF TLQERQMLGL QGLLPPKIET QDIQALRFHR NLKKMTSPLE KYIYIMGIQ ERNEKLFYRI LQDDIESLMP IVYTPTVGLA CSQYGHIFRR PKGLFISISD RGHVRSIVDN WPENHVKAVV V TDGERILG ...String: MLSRLRVVST TCTLACRHLH IKEKGKPLML NPRTNKGMAF TLQERQMLGL QGLLPPKIET QDIQALRFHR NLKKMTSPLE KYIYIMGIQ ERNEKLFYRI LQDDIESLMP IVYTPTVGLA CSQYGHIFRR PKGLFISISD RGHVRSIVDN WPENHVKAVV V TDGERILG LGDLGVYGMG IPVGKLCLYT ACAGIRPDRC LPVCIDVGTD NIALLKDPFY MGLYQKRDRT QQYDDLIDEF MK AITDRYG RNTLIQFEDF GNHNAFRFLR KYREKYCTFN DDIQGTAAVA LAGLLAAQKV ISKPISEHKI LFLGAGEAAL GIA NLIVMS MVENGLSEQE AQKKIWMFDK YGLLVKGRKA KIDSYQEPFT HSAPESIPDT FEDAVNILKP STIIGVAGAG RLFT PDVIR AMASINERPV IFALSNPTAQ AECTAEEAYT LTEGRCLFAS GSPFGPVKLT DGRVFTPGQG NNVYIFPGVA LAVIL CNTR HISDSVFLEA AKALTSQLTD EELAQGRLYP PLANIQEVSI NIAIKVTEYL YANKMAFRYP EPEDKAKYVK ERTWRS EYD SLLPDVYEWP ESASSPPVIT E UniProtKB: NAD-dependent malic enzyme, mitochondrial |
-Macromolecule #2: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 4 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)