[English] 日本語
 Yorodumi
Yorodumi- EMDB-33040: Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / RNA-binding / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology | : / CRISPR type III-associated protein / RAMP superfamily / defense response to virus / RAMP superfamily protein Function and homology information Function and homology information | |||||||||
| Biological species |  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) | |||||||||
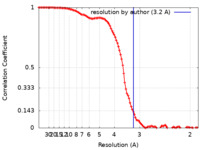
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Yu G / Wang X / Deng Z / Zhang H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2022 Journal: Nat Microbiol / Year: 2022Title: Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex. Authors: Guimei Yu / Xiaoshen Wang / Yi Zhang / Qiyin An / Yanan Wen / Xuzichao Li / Hang Yin / Zengqin Deng / Heng Zhang /  Abstract: The type III-E CRISPR-Cas system uses a single multidomain effector called Cas7-11 (also named gRAMP) to cleave RNA and associate with a caspase-like protease Csx29, showing promising potential for ...The type III-E CRISPR-Cas system uses a single multidomain effector called Cas7-11 (also named gRAMP) to cleave RNA and associate with a caspase-like protease Csx29, showing promising potential for RNA-targeting applications. The structural and molecular mechanisms of the type III-E CRISPR-Cas system remain poorly understood. Here we report four cryo-electron microscopy structures of Cas7-11 at different functional states. Cas7-11 has four Cas7-like domains, which assemble into a helical filament to accommodate CRISPR RNA (crRNA), and a Cas11-like domain facilitating crRNA-target RNA duplex formation. The Cas7.1 domain is critical for crRNA maturation, whereas Cas7.2 and Cas7.3 are responsible for target RNA cleavage. Target RNA binding induces the structural arrangements of Csx29, potentially exposing the catalytic site of Csx29. These results delineate the molecular mechanisms underlying pre-crRNA processing, target RNA recognition and cleavage for Cas7-11, and provide a structural framework to understand the role of Csx29 in type III-E CRISPR system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33040.map.gz emd_33040.map.gz | 94.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33040-v30.xml emd-33040-v30.xml emd-33040.xml emd-33040.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_33040_fsc.xml emd_33040_fsc.xml | 13.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_33040.png emd_33040.png | 111.1 KB | ||
| Filedesc metadata |  emd-33040.cif.gz emd-33040.cif.gz | 7.3 KB | ||
| Others |  emd_33040_half_map_1.map.gz emd_33040_half_map_1.map.gz emd_33040_half_map_2.map.gz emd_33040_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33040 http://ftp.pdbj.org/pub/emdb/structures/EMD-33040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33040 | HTTPS FTP |
-Validation report
| Summary document |  emd_33040_validation.pdf.gz emd_33040_validation.pdf.gz | 880.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33040_full_validation.pdf.gz emd_33040_full_validation.pdf.gz | 879.8 KB | Display | |
| Data in XML |  emd_33040_validation.xml.gz emd_33040_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_33040_validation.cif.gz emd_33040_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33040 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33040 | HTTPS FTP |
-Related structure data
| Related structure data |  7x7aMC  7x7rC  7x8aC  7xc7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33040.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33040.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: unmasked
| File | emd_33040_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
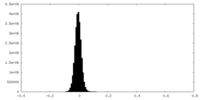
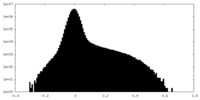
| Density Histograms |
-Half map: unmasked
| File | emd_33040_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unmasked | ||||||||||||
| Projections & Slices |
| ||||||||||||
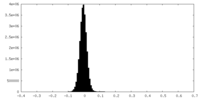
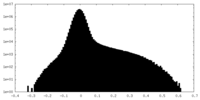
| Density Histograms |
- Sample components
Sample components
-Entire : binary complex
| Entire | Name: binary complex |
|---|---|
| Components |
|
-Supramolecule #1: binary complex
| Supramolecule | Name: binary complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
-Macromolecule #1: RAMP superfamily protein
| Macromolecule | Name: RAMP superfamily protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 197.823797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSNDMNITV ELTFFEPYRL VEWFDWDARK KSHSAMRGQA FAQWTWKGKG RTAGKSFITG TLVRSAVIKA VEELLSLNNG KWEGVPCCN GSFQTDESKG KKPSFLRKRH TLQWQANNKN ICDKEEACPF CILLGRFDNA GKVHERNKDY DIHFSNFDLD H KQEKNDLR ...String: MKSNDMNITV ELTFFEPYRL VEWFDWDARK KSHSAMRGQA FAQWTWKGKG RTAGKSFITG TLVRSAVIKA VEELLSLNNG KWEGVPCCN GSFQTDESKG KKPSFLRKRH TLQWQANNKN ICDKEEACPF CILLGRFDNA GKVHERNKDY DIHFSNFDLD H KQEKNDLR LVDIASGRIL NRVDFDTGKA KDYFRTWEAD YETYGTYTGR ITLRNEHAKK LLLASLGFVD KLCGALCRIE VI KKSESPL PSDTKEQSYT KDDTVEVLSE DHNDELRKQA EVIVEAFKQN DKLEKIRILA DAIRTLRLHG EGVIEKDELP DGK EERDKG HHLWDIKVQG TALRTKLKEL WQSNKDIGWR KFTEMLGSNL YLIYKKETGG VSTRFRILGD TEYYSKAHDS EGSD LFIPV TPPEGIETKE WIIVGRLKAA TPFYFGVQQP SDSIPGKEKK SEDSLVINEH TSFNILLDKE NRYRIPRSAL RGALR RDLR TAFGSGCNVS LGGQILCNCK VCIEMRRITL KDSVSDFSEP PEIRYRIAKN PGTATVEDGS LFDIEVGPEG LTFPFV LRY RGHKFPEQLS SVIRYWEEND GKNGMAWLGG LDSTGKGRFA LKDIKIFEWD LNQKINEYIK ERGMRGKEKE LLEMGES SL PDGLIPYKFF EERECLFPYK ENLKPQWSEV QYTIEVGSPL LTADTISALT EPGNRDAIAY KKRVYNDGNN AIEPEPRF A VKSETHRGIF RTAVGRRTGD LGKEDHEDCT CDMCIIFGNE HESSKIRFED LELINGNEFE KLEKHIDHVA IDRFTGGAL DKAKFDTYPL AGSPKKPLKL KGRFWIKKGF SGDHKLLITT ALSDIRDGLY PLGSKGGVGY GWVAGISIDD NVPDDFKEMI NKTEMPLPE EVEESNNGPI NNDYVHPGHQ SPKQDHKNKN IYYPHYFLDS GSKVYREKDI ITHEEFTEEL LSGKINCKLE T LTPLIIPD TSDENGLKLQ GNKPGHKNYK FFNINGELMI PGSELRGMLR THFEALTKSC FAIFGEDSTL SWRMNADEKD YK IDSNSIR KMESQRNPKY RIPDELQKEL RNSGNGLFNR LYTSERRFWS DVSNKFENSI DYKREILRCA GRPKNYKGGI IRQ RKDSLM AEELKVHRLP LYDNFDIPDS AYKANDHCRK SATCSTSRGC RERFTCGIKV RDKNRVFLNA ANNNRQYLNN IKKS NHDLY LQYLKGEKKI RFNSKVITGS ERSPIDVIAE LNERGRQTGF IKLSGLNNSN KSQGNTGTTF NSGWDRFELN ILLDD LETR PSKSDYPRPR LLFTKDQYEY NITKRCERVF EIDKGNKTGY PVDDQIKKNY EDILDSYDGI KDQEVAERFD TFTRGS KLK VGDLVYFHID GDNKIDSLIP VRISRKCASK TLGGKLDKAL HPCTGLSDGL CPGCHLFGTT DYKGRVKFGF AKYENGP EW LITRGNNPER SLTLGVLESP RPAFSIPDDE SEIPGRKFYL HHNGWRIIRQ KQLEIRETVQ PERNVTTEVM DKGNVFSF D VRFENLREWE LGLLLQSLDP GKNIAHKLGK GKPYGFGSVK IKIDSLHTFK INSNNDKIKR VPQSDIREYI NKGYQKLIE WSGNNSIQKG NVLPQWHVIP HIDKLYKLLW VPFLNDSKLE PDVRYPVLNE ESKGYIEGSD YTYKKLGDKD NLPYKTRVKG LTTPWSPWN PFQVIAEHEE QEVNVTGSRP SVTDKIERDG KMV UniProtKB: RAMP superfamily protein |
-Macromolecule #2: RNA (33-MER)
| Macromolecule | Name: RNA (33-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Candidatus Scalindua brodae (bacteria) Candidatus Scalindua brodae (bacteria) |
| Molecular weight | Theoretical: 10.404171 KDa |
| Sequence | String: GACUUAAUGU CACGGUACCC AAUUUUCUGC CCC |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
| Details | No further treatment. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)