[English] 日本語
 Yorodumi
Yorodumi- EMDB-33026: lymphocytic choriomeningitis virus RNA-dependent RNA polymerase (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | lymphocytic choriomeningitis virus RNA-dependent RNA polymerase (LCMV-L protein) | |||||||||
 Map data Map data | EM map of LCMV L protein | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | L protein / monomer / RdRp / NSFERASE / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative stranded viral RNA replication / cap snatching / virion component / host cell / Hydrolases; Acting on ester bonds / host cell cytoplasm / hydrolase activity / RNA-directed RNA polymerase / nucleotide binding / RNA-directed RNA polymerase activity / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Lymphocytic choriomeningitis virus (strain Armstrong) Lymphocytic choriomeningitis virus (strain Armstrong) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Liu L / Lou Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2023 Journal: Protein Cell / Year: 2023Title: Structure basis for allosteric regulation of lymphocytic choriomeningitis virus polymerase function by Z matrix protein. Authors: Lu Liu / Panpan Wang / Aijun Liu / Leike Zhang / Liming Yan / Yu Guo / Gengfu Xiao / Zihe Rao / Zhiyong Lou /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33026.map.gz emd_33026.map.gz | 28.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33026-v30.xml emd-33026-v30.xml emd-33026.xml emd-33026.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33026.png emd_33026.png | 74 KB | ||
| Filedesc metadata |  emd-33026.cif.gz emd-33026.cif.gz | 7.2 KB | ||
| Others |  emd_33026_half_map_1.map.gz emd_33026_half_map_1.map.gz emd_33026_half_map_2.map.gz emd_33026_half_map_2.map.gz | 28.3 MB 28.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33026 http://ftp.pdbj.org/pub/emdb/structures/EMD-33026 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33026 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33026 | HTTPS FTP |
-Validation report
| Summary document |  emd_33026_validation.pdf.gz emd_33026_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33026_full_validation.pdf.gz emd_33026_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_33026_validation.xml.gz emd_33026_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_33026_validation.cif.gz emd_33026_validation.cif.gz | 12.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33026 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33026 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33026 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33026 | HTTPS FTP |
-Related structure data
| Related structure data |  7x6sMC  7x6vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_33026.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33026.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of LCMV L protein | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||
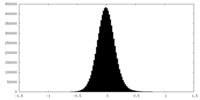
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33026_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
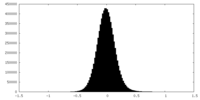
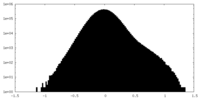
| Density Histograms |
-Half map: #1
| File | emd_33026_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
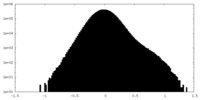
| Density Histograms |
- Sample components
Sample components
-Entire : Lymphocytic choriomeningitis virus polymerase L protein
| Entire | Name: Lymphocytic choriomeningitis virus polymerase L protein |
|---|---|
| Components |
|
-Supramolecule #1: Lymphocytic choriomeningitis virus polymerase L protein
| Supramolecule | Name: Lymphocytic choriomeningitis virus polymerase L protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Lymphocytic choriomeningitis virus (strain Armstrong) Lymphocytic choriomeningitis virus (strain Armstrong) |
-Macromolecule #1: RNA-directed RNA polymerase L
| Macromolecule | Name: RNA-directed RNA polymerase L / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lymphocytic choriomeningitis virus (strain Armstrong) Lymphocytic choriomeningitis virus (strain Armstrong)Strain: Armstrong |
| Molecular weight | Theoretical: 254.894641 KDa |
| Recombinant expression | Organism:  unidentified baculovirus unidentified baculovirus |
| Sequence | String: MDEIISELRE LCLNYIEQDE RLSRQKLNFL GQREPRMVLI EGLKLLSRCI EIDSADKSGC THNHDDKSVE TILVESGIVC PGLPLIIPD GYKLIDNSLI LLECFVRSTP ASFEKKFIED TNKLACIRED LAVAGVTLVP IVDGRCDYDN SFMPEWANFK F RDLLFKLL ...String: MDEIISELRE LCLNYIEQDE RLSRQKLNFL GQREPRMVLI EGLKLLSRCI EIDSADKSGC THNHDDKSVE TILVESGIVC PGLPLIIPD GYKLIDNSLI LLECFVRSTP ASFEKKFIED TNKLACIRED LAVAGVTLVP IVDGRCDYDN SFMPEWANFK F RDLLFKLL EYSNQNEKVF EESEYFRLCE SLKTTIDKRS GMDSMKILKD ARSTHNDEIM RMCHEGINPN MSCDDVVFGI NS LFSRFRR DLESGKLKRN FQKVNPEGLI KEFSELYENL ADSDDILTLS REAVESCPLM RFITAETHGH ERGSETSTEY ERL LSMLNK VKSLKLLNTR RRQLLNLDVL CLSSLIKQSK FKGLKNDKHW VGCCYSSVND RLVSFHSTKE EFIRLLRNRK KSKV FRKVS FEELFRASIS EFIAKIQKCL LVVGLSFEHY GLSEHLEQEC HIPFTEFENF MKIGAHPIMY YTKFEDYNFQ PSTEQ LKNI QSLRRLSSVC LALTNSMKTS SVARLRQNQI GSVRYQVVEC KEVFCQVIKL DSEEYHLLYQ KTGESSRCYS IQGPDG HLI SFYADPKRFF LPIFSDEVLY NMIDIMISWI RSCPDLKDCL TDIEVALRTL LLLMLTNPTK RNQKQVQSVR YLVMAIV SD FSSTSLMDKL REDLITPAEK VVYKLLRFLI KTIFGTGEKV LLSAKFKFML NVSYLCHLIT KETPDRLTDQ IKCFEKFF E PKSQFGFFVN PKEAITPEEE CVFYEQMKRF TSKEIDCQHT TPGVNLEAFS LMVSSFNNGT LIFKGEKKLN SLDPMTNSG CATALDLASN KSVVVNKHLN GERLLEYDFN KLLVSAVSQI TESFVRKQKY KLSHSDYEYK VSKLVSRLVI GSKGEETGRS EDNLAEICF DGEEETSFFK SLEEKVNTTI ARYRRGRRAN DKGDGEKLTN TKGLHHLQLI LTGKMAHLRK VILSEISFHL V EDFDPSCL TNDDMKFICE AVEGSTELSP LYFTSVIKDQ CGLDEMAKNL CRKFFSENDW FSCMKMILLQ MNANAYSGKY RH MQRQGLN FKFDWDKLEE DVRISERESN SESLSKALSL TKCMSAALKN LCFYSEESPT SYTSVGPDSG RLKFALSYKE QVG GNRELY IGDLRTKMFT RLIEDYFESF SSFFSGSCLN NDKEFENAIL SMTINVREGF LNYSMDHSKW GPMMCPFLFL MFLQ NLKLG DDQYVRSGKD HVSTLLTWHM HKLVEVPFPV VNAMMKSYVK SKLKLLRGSE TTVTERIFRQ YFEMGIVPSH ISSLI DMGQ GILHNASDFY GLLSERFINY CIGVIFGERP EAYTSSDDQI TLFDRRLSDL VVSDPEEVLV LLEFQSHLSG LLNKFI SPK SVAGRFAAEF KSRFYVWGEE VPLLTKFVSA ALHNVKCKEP HQLCETIDTI ADQAIANGVP VSLVNSIQRR TLDLLKY AN FPLDPFLLNT NTDVKDWLDG SRGYRIQRLI EELCPNETKV VRKLVRKLHH KLKNGEFNEE FFLDLFNRDK KEAILQLG D LLGLEEDLNQ LADVNWLNLN EMFPLRMVLR QKVVYPSVMT FQEERIPSLI KTLQNKLCSK FTRGAQKLLS EAINKSAFQ SCISSGFIGL CKTLGSRCVR NKNRENLYIK KLLEDLTTDD HVTRVCNRDG ITLYICDKQS HPEAHRDHIC LLRPLLWDYI CISLSNSFE LGVWVLAEPT KGKNNSENLT LKHLNPCDYV ARKPESSRLL EDKVNLNQVI QSVRRLYPKI FEDQLLPFMS D MSSKNMRW SPRIKFLDLC VLIDINSESL SLISHVVKWK RDEHYTVLFS DLANSHQRSD SSLVDEFVVS TRDVCKNFLK QV YFESFVR EFVATTRTLG NFSWFPHKEM MPSEDGAEAL GPFQSFVSKV VNKNVERPMF RNDLQFGFGW FSYRMGDVVC NAA MLIRQG LTNPKAFKSL KDLWDYMLNY TKGVLEFSIS VDFTHNQNNT DCLRKFSLIF LVRCQLQNPG VAELLSCSHL FKGE IDRRM LDECLHLLRT DSVFKVNDGV FDIRSEEFED YMEDPLILGD SLELELLGSK RILDGIRSID FERVGPEWEP VPLTV KMGA LFEGRNLVQN IIVKLETKDM KVFLAGLEGY EKISDVLGNL FLHRFRTGEH LLGSEISVIL QELCIDRSIL LIPLSL LPD WFAFKDCRLC FSKSRSTLMY ETVGGRFRLK GRSCDDWLGG SVAEDID UniProtKB: RNA-directed RNA polymerase L |
-Macromolecule #2: MANGANESE (II) ION
| Macromolecule | Name: MANGANESE (II) ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: MN |
|---|---|
| Molecular weight | Theoretical: 54.938 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)