+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of human TRiC-ATP-closed state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationzona pellucida receptor complex / positive regulation of establishment of protein localization to telomere / positive regulation of protein localization to Cajal body / scaRNA localization to Cajal body / positive regulation of telomerase RNA localization to Cajal body / chaperonin-containing T-complex / BBSome-mediated cargo-targeting to cilium / tubulin complex assembly / sperm head-tail coupling apparatus / Formation of tubulin folding intermediates by CCT/TriC ...zona pellucida receptor complex / positive regulation of establishment of protein localization to telomere / positive regulation of protein localization to Cajal body / scaRNA localization to Cajal body / positive regulation of telomerase RNA localization to Cajal body / chaperonin-containing T-complex / BBSome-mediated cargo-targeting to cilium / tubulin complex assembly / sperm head-tail coupling apparatus / Formation of tubulin folding intermediates by CCT/TriC / binding of sperm to zona pellucida / Folding of actin by CCT/TriC / Prefoldin mediated transfer of substrate to CCT/TriC / RHOBTB1 GTPase cycle / WD40-repeat domain binding / Association of TriC/CCT with target proteins during biosynthesis / pericentriolar material / Hydrolases; Acting on acid anhydrides; In phosphorus-containing anhydrides / chaperone-mediated protein complex assembly / RHOBTB2 GTPase cycle / beta-tubulin binding / heterochromatin / positive regulation of telomere maintenance via telomerase / protein folding chaperone / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / acrosomal vesicle / mRNA 3'-UTR binding / ATP-dependent protein folding chaperone / mRNA 5'-UTR binding / response to virus / azurophil granule lumen / melanosome / unfolded protein binding / sperm midpiece / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / G-protein beta-subunit binding / protein folding / cell body / secretory granule lumen / ficolin-1-rich granule lumen / microtubule / cytoskeleton / protein stabilization / cadherin binding / ubiquitin protein ligase binding / Neutrophil degranulation / centrosome / Golgi apparatus / ATP hydrolysis activity / RNA binding / extracellular exosome / extracellular region / nucleoplasm / ATP binding / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Cong Y / Liu CX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM. Authors: Caixuan Liu / Mingliang Jin / Shutian Wang / Wenyu Han / Qiaoyu Zhao / Yifan Wang / Cong Xu / Lei Diao / Yue Yin / Chao Peng / Lan Bao / Yanxing Wang / Yao Cong /  Abstract: The eukaryotic chaperonin TRiC/CCT assists the folding of about 10% of cytosolic proteins through an ATP-driven conformational cycle, and the essential cytoskeleton protein tubulin is the obligate ...The eukaryotic chaperonin TRiC/CCT assists the folding of about 10% of cytosolic proteins through an ATP-driven conformational cycle, and the essential cytoskeleton protein tubulin is the obligate substrate of TRiC. Here, we present an ensemble of cryo-EM structures of endogenous human TRiC throughout its ATPase cycle, with three of them revealing endogenously engaged tubulin in different folding stages. The open-state TRiC-tubulin-S1 and -S2 maps show extra density corresponding to tubulin in the cis-ring chamber of TRiC. Our structural and XL-MS analyses suggest a gradual upward translocation and stabilization of tubulin within the TRiC chamber accompanying TRiC ring closure. In the closed TRiC-tubulin-S3 map, we capture a near-natively folded tubulin-with the tubulin engaging through its N and C domains mainly with the A and I domains of the CCT3/6/8 subunits through electrostatic and hydrophilic interactions. Moreover, we also show the potential role of TRiC C-terminal tails in substrate stabilization and folding. Our study delineates the pathway and molecular mechanism of TRiC-mediated folding of tubulin along the ATPase cycle of TRiC, and may also inform the design of therapeutic agents targeting TRiC-tubulin interactions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33025.map.gz emd_33025.map.gz | 17.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33025-v30.xml emd-33025-v30.xml emd-33025.xml emd-33025.xml | 26.9 KB 26.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33025.png emd_33025.png | 210.3 KB | ||
| Filedesc metadata |  emd-33025.cif.gz emd-33025.cif.gz | 8.4 KB | ||
| Others |  emd_33025_additional_1.map.gz emd_33025_additional_1.map.gz emd_33025_half_map_1.map.gz emd_33025_half_map_1.map.gz emd_33025_half_map_2.map.gz emd_33025_half_map_2.map.gz | 17.4 MB 140.6 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33025 http://ftp.pdbj.org/pub/emdb/structures/EMD-33025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33025 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33025 | HTTPS FTP |
-Related structure data
| Related structure data |  7x6qMC  7wz3C  7x0aC  7x0sC  7x0vC  7x3jC  7x3uC  7x7yC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33025.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33025.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_33025_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
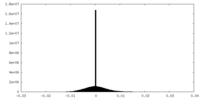
| Density Histograms |
-Half map: #1
| File | emd_33025_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
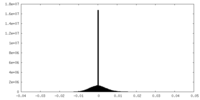
| Density Histograms |
-Half map: #2
| File | emd_33025_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
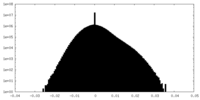
| Density Histograms |
- Sample components
Sample components
-Entire : Human TRiC-ATP-closed state
| Entire | Name: Human TRiC-ATP-closed state |
|---|---|
| Components |
|
-Supramolecule #1: Human TRiC-ATP-closed state
| Supramolecule | Name: Human TRiC-ATP-closed state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: T-complex protein 1 subunit alpha
| Macromolecule | Name: T-complex protein 1 subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.418477 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEGPLSVFGD RSTGETIRSQ NVMAAASIAN IVKSSLGPVG LDKMLVDDIG DVTITNDGAT ILKLLEVEHP AAKVLCELAD LQDKEVGDG TTSVVIIAAE LLKNADELVK QKIHPTSVIS GYRLACKEAV RYINENLIVN TDELGRDCLI NAAKTSMSSK I IGINGDFF ...String: MEGPLSVFGD RSTGETIRSQ NVMAAASIAN IVKSSLGPVG LDKMLVDDIG DVTITNDGAT ILKLLEVEHP AAKVLCELAD LQDKEVGDG TTSVVIIAAE LLKNADELVK QKIHPTSVIS GYRLACKEAV RYINENLIVN TDELGRDCLI NAAKTSMSSK I IGINGDFF ANMVVDAVLA IKYTDIRGQP RYPVNSVNIL KAHGRSQMES MLISGYALNC VVGSQGMPKR IVNAKIACLD FS LQKTKMK LGVQVVITDP EKLDQIRQRE SDITKERIQK ILATGANVIL TTGGIDDMCL KYFVEAGAMA VRRVLKRDLK RIA KASGAT ILSTLANLEG EETFEAAMLG QAEEVVQERI CDDELILIKN TKARTSASII LRGANDFMCD EMERSLHDAL CVVK RVLES KSVVPGGGAV EAALSIYLEN YATSMGSREQ LAIAEFARSL LVIPNTLAVN AAQDSTDLVA KLRAFHNEAQ VNPER KNLK WIGLDLSNGK PRDNKQAGVF EPTIVKVKSL KFATEAAITI LRIDDLIKLH PESKDDKHGS YEDAVHSGAL ND UniProtKB: T-complex protein 1 subunit alpha |
-Macromolecule #2: T-complex protein 1 subunit beta
| Macromolecule | Name: T-complex protein 1 subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.567141 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MASLSLAPVN IFKAGADEER AETARLTSFI GAIAIGDLVK STLGPKGMDK ILLSSGRDAS LMVTNDGATI LKNIGVDNPA AKVLVDMSR VQDDEVGDGT TSVTVLAAEL LREAESLIAK KIHPQTIIAG WREATKAARE ALLSSAVDHG SDEVKFRQDL M NIAGTTLS ...String: MASLSLAPVN IFKAGADEER AETARLTSFI GAIAIGDLVK STLGPKGMDK ILLSSGRDAS LMVTNDGATI LKNIGVDNPA AKVLVDMSR VQDDEVGDGT TSVTVLAAEL LREAESLIAK KIHPQTIIAG WREATKAARE ALLSSAVDHG SDEVKFRQDL M NIAGTTLS SKLLTHHKDH FTKLAVEAVL RLKGSGNLEA IHIIKKLGGS LADSYLDEGF LLDKKIGVNQ PKRIENAKIL IA NTGMDTD KIKIFGSRVR VDSTAKVAEI EHAEKEKMKE KVERILKHGI NCFINRQLIY NYPEQLFGAA GVMAIEHADF AGV ERLALV TGGEIASTFD HPELVKLGSC KLIEEVMIGE DKLIHFSGVA LGEACTIVLR GATQQILDEA ERSLHDALCV LAQT VKDSR TVYGGGCSEM LMAHAVTQLA NRTPGKEAVA MESYAKALRM LPTIIADNAG YDSADLVAQL RAAHSEGNTT AGLDM REGT IGDMAILGIT ESFQVKRQVL LSAAEAAEVI LRVDNIIKAA PRKRVPDHHP C UniProtKB: T-complex protein 1 subunit beta |
-Macromolecule #3: T-complex protein 1 subunit epsilon
| Macromolecule | Name: T-complex protein 1 subunit epsilon / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.547684 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SMGTLAFDEY GRPFLIIKDQ DRKSRLMGLE ALKSHIMAAK AVANTMRTSL GPNGLDKMMV DKDGDVTVTN DGATILSMMD VDHQIAKLM VELSKSQDDE IGDGTTGVVV LAGALLEEAE QLLDRGIHPI RIADGYEQAA RVAIEHLDKI SDSVLVDIKD T EPLIQTAK ...String: SMGTLAFDEY GRPFLIIKDQ DRKSRLMGLE ALKSHIMAAK AVANTMRTSL GPNGLDKMMV DKDGDVTVTN DGATILSMMD VDHQIAKLM VELSKSQDDE IGDGTTGVVV LAGALLEEAE QLLDRGIHPI RIADGYEQAA RVAIEHLDKI SDSVLVDIKD T EPLIQTAK TTLGSKVVNS CHRQMAEIAV NAVLTVADME RRDVDFELIK VEGKVGGRLE DTKLIKGVIV DKDFSHPQMP KK VEDAKIA ILTCPFEPPK PKTKHKLDVT SVEDYKALQK YEKEKFEEMI QQIKETGANL AICQWGFDDE ANHLLLQNNL PAV RWVGGP EIELIAIATG GRIVPRFSEL TAEKLGFAGL VQEISFGTTK DKMLVIEQCK NSRAVTIFIR GGNKMIIEEA KRSL HDALC VIRNLIRDNR VVYGGGAAEI SCALAVSQEA DKCPTLEQYA MRAFADALEV IPMALSENSG MNPIQTMTEV RARQV KEMN PALGIDCLHK GTNDMKQQHV IETLIGKKQQ ISLATQMVRM ILKIDDIRKP GESEE UniProtKB: T-complex protein 1 subunit epsilon |
-Macromolecule #4: T-complex protein 1 subunit gamma
| Macromolecule | Name: T-complex protein 1 subunit gamma / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.613855 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMGHRPVLVL SQNTKRESGR KVQSGNINAA KTIADIIRTC LGPKSMMKML LDPMGGIVMT NDGNAILREI QVQHPAAKSM IEISRTQDE EVGDGTTSVI ILAGEMLSVA EHFLEQQMHP TVVISAYRKA LDDMISTLKK ISIPVDISDS DMMLNIINSS I TTKAISRW ...String: MMGHRPVLVL SQNTKRESGR KVQSGNINAA KTIADIIRTC LGPKSMMKML LDPMGGIVMT NDGNAILREI QVQHPAAKSM IEISRTQDE EVGDGTTSVI ILAGEMLSVA EHFLEQQMHP TVVISAYRKA LDDMISTLKK ISIPVDISDS DMMLNIINSS I TTKAISRW SSLACNIALD AVKMVQFEEN GRKEIDIKKY ARVEKIPGGI IEDSCVLRGV MINKDVTHPR MRRYIKNPRI VL LDSSLEY KKGESQTDIE ITREEDFTRI LQMEEEYIQQ LCEDIIQLKP DVVITEKGIS DLAQHYLMRA NITAIRRVRK TDN NRIARA CGARIVSRPE ELREDDVGTG AGLLEIKKIG DEYFTFITDC KDPKACTILL RGASKEILSE VERNLQDAMQ VCRN VLLDP QLVPGGGASE MAVAHALTEK SKAMTGVEQW PYRAVAQALE VIPRTLIQNC GASTIRLLTS LRAKHTQENC ETWGV NGET GTLVDMKELG IWEPLAVKLQ TYKTAVETAV LLLRIDDIVS GHKKKGDDQS RQGGAPDAGQ E UniProtKB: T-complex protein 1 subunit gamma |
-Macromolecule #5: T-complex protein 1 subunit eta
| Macromolecule | Name: T-complex protein 1 subunit eta / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.417457 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MMPTPVILLK EGTDSSQGIP QLVSNISACQ VIAEAVRTTL GPRGMDKLIV DGRGKATISN DGATILKLLD VVHPAAKTLV DIAKSQDAE VGDGTTSVTL LAAEFLKQVK PYVEEGLHPQ IIIRAFRTAT QLAVNKIKEI AVTVKKADKV EQRKLLEKCA M TALSSKLI ...String: MMPTPVILLK EGTDSSQGIP QLVSNISACQ VIAEAVRTTL GPRGMDKLIV DGRGKATISN DGATILKLLD VVHPAAKTLV DIAKSQDAE VGDGTTSVTL LAAEFLKQVK PYVEEGLHPQ IIIRAFRTAT QLAVNKIKEI AVTVKKADKV EQRKLLEKCA M TALSSKLI SQQKAFFAKM VVDAVMMLDD LLQLKMIGIK KVQGGALEDS QLVAGVAFKK TFSYAGFEMQ PKKYHNPKIA LL NVELELK AEKDNAEIRV HTVEDYQAIV DAEWNILYDK LEKIHHSGAK VVSSKLPIGD VATQYFADRD MFCAGRVPEE DLK RTMMAC GGSIQTSVNA LSADVLGRCQ VFEETQIGGE RYNFFTGCPK AKTCTFILRG GAEQFMEETE RSLHDAIMIV RRAI KNDSV VAGGGAIEME LSKYLRDYSR TIPGKQQLLI GAYAKALEII PRQLCDNAGF DATNILNKLR ARHAQGGTWY GVDIN NEDI ADNFEAFVWE PAMVRINALT AASEAACLIV SVDETIKNPR STVDAPTAAG RGRGRGRPH UniProtKB: T-complex protein 1 subunit eta |
-Macromolecule #6: T-complex protein 1 subunit delta
| Macromolecule | Name: T-complex protein 1 subunit delta / type: protein_or_peptide / ID: 6 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.996113 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPENVAPRSG ATAGAAGGRG KGAYQDRDKP AQIRFSNISA AKAVADAIRT SLGPKGMDKM IQDGKGDVTI TNDGATILKQ MQVLHPAAR MLVELSKAQD IEAGDGTTSV VIIAGSLLDS CTKLLQKGIH PTIISESFQK ALEKGIEILT DMSRPVELSD R ETLLNSAT ...String: MPENVAPRSG ATAGAAGGRG KGAYQDRDKP AQIRFSNISA AKAVADAIRT SLGPKGMDKM IQDGKGDVTI TNDGATILKQ MQVLHPAAR MLVELSKAQD IEAGDGTTSV VIIAGSLLDS CTKLLQKGIH PTIISESFQK ALEKGIEILT DMSRPVELSD R ETLLNSAT TSLNSKVVSQ YSSLLSPMSV NAVMKVIDPA TATSVDLRDI KIVKKLGGTI DDCELVEGLV LTQKVSNSGI TR VEKAKIG LIQFCLSAPK TDMDNQIVVS DYAQMDRVLR EERAYILNLV KQIKKTGCNV LLIQKSILRD ALSDLALHFL NKM KIMVIK DIEREDIEFI CKTIGTKPVA HIDQFTADML GSAELAEEVN LNGSGKLLKI TGCASPGKTV TIVVRGSNKL VIEE AERSI HDALCVIRCL VKKRALIAGG GAPEIELALR LTEYSRTLSG MESYCVRAFA DAMEVIPSTL AENAGLNPIS TVTEL RNRH AQGEKTAGIN VRKGGISNIL EELVVQPLLV SVSALTLATE TVRSILKIDD VVNTR UniProtKB: T-complex protein 1 subunit delta |
-Macromolecule #7: T-complex protein 1 subunit theta
| Macromolecule | Name: T-complex protein 1 subunit theta / type: protein_or_peptide / ID: 7 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.691422 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MALHVPKAPG FAQMLKEGAK HFSGLEEAVY RNIQACKELA QTTRTAYGPN GMNKMVINHL EKLFVTNDAA TILRELEVQH PAAKMIVMA SHMQEQEVGD GTNFVLVFAG ALLELAEELL RIGLSVSEVI EGYEIACRKA HEILPNLVCC SAKNLRDIDE V SSLLRTSI ...String: MALHVPKAPG FAQMLKEGAK HFSGLEEAVY RNIQACKELA QTTRTAYGPN GMNKMVINHL EKLFVTNDAA TILRELEVQH PAAKMIVMA SHMQEQEVGD GTNFVLVFAG ALLELAEELL RIGLSVSEVI EGYEIACRKA HEILPNLVCC SAKNLRDIDE V SSLLRTSI MSKQYGNEVF LAKLIAQACV SIFPDSGHFN VDNIRVCKIL GSGISSSSVL HGMVFKKETE GDVTSVKDAK IA VYSCPFD GMITETKGTV LIKTAEELMN FSKGEENLMD AQVKAIADTG ANVVVTGGKV ADMALHYANK YNIMLVRLNS KWD LRRLCK TVGATALPRL TPPVLEEMGH CDSVYLSEVG DTQVVVFKHE KEDGAISTIV LRGSTDNLMD DIERAVDDGV NTFK VLTRD KRLVPGGGAT EIELAKQITS YGETCPGLEQ YAIKKFAEAF EAIPRALAEN SGVKANEVIS KLYAVHQEGN KNVGL DIEA EVPAVKDMLE AGILDTYLGK YWAIKLATNA AVTVLRVDQI IMAKPAGGPK PPSGKKDWDD DQND UniProtKB: T-complex protein 1 subunit theta |
-Macromolecule #8: T-complex protein 1 subunit zeta
| Macromolecule | Name: T-complex protein 1 subunit zeta / type: protein_or_peptide / ID: 8 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.106086 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAAVKTLNPK AEVARAQAAL AVNISAARGL QDVLRTNLGP KGTMKMLVSG AGDIKLTKDG NVLLHEMQIQ HPTASLIAKV ATAQDDITG DGTTSNVLII GELLKQADLY ISEGLHPRII TEGFEAAKEK ALQFLEEVKV SREMDRETLI DVARTSLRTK V HAELADVL ...String: MAAVKTLNPK AEVARAQAAL AVNISAARGL QDVLRTNLGP KGTMKMLVSG AGDIKLTKDG NVLLHEMQIQ HPTASLIAKV ATAQDDITG DGTTSNVLII GELLKQADLY ISEGLHPRII TEGFEAAKEK ALQFLEEVKV SREMDRETLI DVARTSLRTK V HAELADVL TEAVVDSILA IKKQDEPIDL FMIEIMEMKH KSETDTSLIR GLVLDHGARH PDMKKRVEDA YILTCNVSLE YE KTEVNSG FFYKSAEERE KLVKAERKFI EDRVKKIIEL KRKVCGDSDK GFVVINQKGI DPFSLDALSK EGIVALRRAK RRN MERLTL ACGGVALNSF DDLSPDCLGH AGLVYEYTLG EEKFTFIEKC NNPRSVTLLI KGPNKHTLTQ IKDAVRDGLR AVKN AIDDG CVVPGAGAVE VAMAEALIKH KPSVKGRAQL GVQAFADALL IIPKVLAQNS GFDLQETLVK IQAEHSESGQ LVGVD LNTG EPMVAAEVGV WDNYCVKKQL LHSCTVIATN ILLVDEIMRA GMSSLKG UniProtKB: T-complex protein 1 subunit zeta |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Component - Concentration: 50.0 mM / Component - Formula: NaCl / Component - Name: sodium Chloride |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 57.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 41646 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)