[English] 日本語
 Yorodumi
Yorodumi- EMDB-33023: Cryo-EM structure of H3 hemagglutinin from A/HongKong/01/1968 in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of H3 hemagglutinin from A/HongKong/01/1968 in complex with a neutralizing antibody 28-12 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | H3 hemagglutinin / A/HongKong/01/1968 / antibody 28-12 / cryo-EM / STRUCTURAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |   Influenza A virus / Influenza A virus /  Homo sapiens (human) / Homo sapiens (human) /  Influenza A virus (A/Hong Kong/1/1968(H3N2)) Influenza A virus (A/Hong Kong/1/1968(H3N2)) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Cong Y / Liu CX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Unique binding pattern for a lineage of human antibodies with broad reactivity against influenza A virus. Authors: Xiaoyu Sun / Caixuan Liu / Xiao Lu / Zhiyang Ling / Chunyan Yi / Zhen Zhang / Zi Li / Mingliang Jin / Wenshuai Wang / Shubing Tang / Fangfang Wang / Fang Wang / Sonam Wangmo / Shuangfeng ...Authors: Xiaoyu Sun / Caixuan Liu / Xiao Lu / Zhiyang Ling / Chunyan Yi / Zhen Zhang / Zi Li / Mingliang Jin / Wenshuai Wang / Shubing Tang / Fangfang Wang / Fang Wang / Sonam Wangmo / Shuangfeng Chen / Li Li / Liyan Ma / Yaguang Zhang / Zhuo Yang / Xiaoping Dong / Zhikang Qian / Jianping Ding / Dayan Wang / Yao Cong / Bing Sun /  Abstract: Most structurally characterized broadly neutralizing antibodies (bnAbs) against influenza A viruses (IAVs) target the conserved conformational epitopes of hemagglutinin (HA). Here, we report a ...Most structurally characterized broadly neutralizing antibodies (bnAbs) against influenza A viruses (IAVs) target the conserved conformational epitopes of hemagglutinin (HA). Here, we report a lineage of naturally occurring human antibodies sharing the same germline gene, V3-48/V1-12. These antibodies broadly neutralize the major circulating strains of IAV in vitro and in vivo mainly by binding a contiguous epitope of H3N2 HA, but a conformational epitope of H1N1 HA, respectively. Our structural and functional studies of antibody 28-12 revealed that the continuous amino acids in helix A, particularly N49 of H3 HA, are critical to determine the binding feature with 28-12. In contrast, the conformational epitope feature is dependent on the discontinuous segments involving helix A, the fusion peptide, and several HA1 residues within H1N1 HA. We report that this antibody was initially selected by H3 (group 2) viruses and evolved via somatic hypermutation to enhance the reactivity to H3 and acquire cross-neutralization to H1 (group 1) virus. These findings enrich our understanding of different antigenic determinants of heterosubtypic influenza viruses for the recognition of bnAbs and provide a reference for the design of influenza vaccines and more effective antiviral drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33023.map.gz emd_33023.map.gz | 36.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33023-v30.xml emd-33023-v30.xml emd-33023.xml emd-33023.xml | 14 KB 14 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33023.png emd_33023.png | 83.4 KB | ||
| Filedesc metadata |  emd-33023.cif.gz emd-33023.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33023 http://ftp.pdbj.org/pub/emdb/structures/EMD-33023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33023 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33023 | HTTPS FTP |
-Related structure data
| Related structure data |  7x6lMC  7x6oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33023.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33023.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.318 Å | ||||||||||||||||||||||||||||||||||||
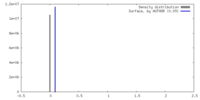
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM structure of H3 hemagglutinin from A/HongKong/01/1968 in ...
| Entire | Name: Cryo-EM structure of H3 hemagglutinin from A/HongKong/01/1968 in complex with a neutralizing antibody 28-12 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of H3 hemagglutinin from A/HongKong/01/1968 in ...
| Supramolecule | Name: Cryo-EM structure of H3 hemagglutinin from A/HongKong/01/1968 in complex with a neutralizing antibody 28-12 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
-Macromolecule #1: Heavy chain of antibody 12 fab
| Macromolecule | Name: Heavy chain of antibody 12 fab / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.466277 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVQPGGSLRL SCAASGFTFS TYNMNWVRQA PGKGLEWLSY ISTSSNTIYY ADSVKGRFTI SRDNAKNSLF LQMNSLRDE DTAVYYCARD RGCSSTNCYV VGYYFYGMDV WGQGTTVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT ...String: EVQLVESGGG LVQPGGSLRL SCAASGFTFS TYNMNWVRQA PGKGLEWLSY ISTSSNTIYY ADSVKGRFTI SRDNAKNSLF LQMNSLRDE DTAVYYCARD RGCSSTNCYV VGYYFYGMDV WGQGTTVTVS SASTKGPSVF PLAPSSKSTS GGTAALGCLV K DYFPEPVT VSWNSGALTS GVHTFPAVLQ SSGLYSLSSV VTVPSSSLGT QTYICNVNHK PSNTKVDKKV E |
-Macromolecule #2: The light chain of the antibody 12 fab
| Macromolecule | Name: The light chain of the antibody 12 fab / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.264861 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIQMTQSPSS VSASVGDRVT ITCRASQGIS SYLAWYQLKP GRAPKLLIYG ATRLQSGVPS RFSGSGSGTD FTLTISGLQP EDFATYHCQ QADSFPLTFG QGTRLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSS VSASVGDRVT ITCRASQGIS SYLAWYQLKP GRAPKLLIYG ATRLQSGVPS RFSGSGSGTD FTLTISGLQP EDFATYHCQ QADSFPLTFG QGTRLEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #3: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/Hong Kong/1/1968(H3N2)) / Strain: A/Hong Kong/1/1968 H3N2 Influenza A virus (A/Hong Kong/1/1968(H3N2)) / Strain: A/Hong Kong/1/1968 H3N2 |
| Molecular weight | Theoretical: 36.218664 KDa |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
| Sequence | String: QDLPGNDNPT ATLCLGHHAV PNGTLVKTIT DDQIEVTNAT ELVQSSSTGK ICNNPHRILD GIDCTLIDAL LGDPHCDVFQ NETWDLFVE RSKAFSNCYP YDVPDYASLR SLVASSGTLE FITEGFTWTG VTQNGGSNAC KRGPGSGFFS RLNWLTKSGS T YPVLNVTM ...String: QDLPGNDNPT ATLCLGHHAV PNGTLVKTIT DDQIEVTNAT ELVQSSSTGK ICNNPHRILD GIDCTLIDAL LGDPHCDVFQ NETWDLFVE RSKAFSNCYP YDVPDYASLR SLVASSGTLE FITEGFTWTG VTQNGGSNAC KRGPGSGFFS RLNWLTKSGS T YPVLNVTM PNNDNFDKLY IWGVHHPSTN QEQTSLYVQA SGRVTVSTRR SQQTIIPNIG SRPWVRGLSS RISIYWTIVK PG DVLVINS NGNLIAPRGY FKMRTGKSSI MRSDAPIDTC ISECITPNGS IPNDKPFQNV NKITYGACPK YVKQNTLKLA TGM RNVPEK QTR UniProtKB: Hemagglutinin |
-Macromolecule #4: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 20.297455 KDa |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
| Sequence | String: GLFGAIAGFI ENGWEGMIDG WYGFRHQNSE GTGQAADLKS TQAAIDQING KLNRVIEKTN EKFHQIEKEF SEVEGRIQDL EKYVEDTKI DLWSYNAELL VALENQHTID LTDSEMNKLF EKTRRQLREN AEDMGNGCFK IYHKCDNACI ESIRNGTYDH D VYRDEALN NRFQIKGV UniProtKB: Hemagglutinin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 38.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 105383 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


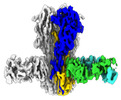







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)