[English] 日本語
 Yorodumi
Yorodumi- EMDB-32918: CryoEM structure of chitin synthase 1 from Phytophthora sojae com... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with UDP | ||||||||||||
 Map data Map data | CryoEM map of chitin synthase 1 from Phytophthora sojae complexed with UDP | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | carbohydate / biosynthetic protein / membrane protein / transferase | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationchitin synthase / chitin synthase activity / chitin biosynthetic process / cell wall organization / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Phytophthora sojae strain P6497 (eukaryote) Phytophthora sojae strain P6497 (eukaryote) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Chen W / Cao P | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Structural basis for directional chitin biosynthesis. Authors: Wei Chen / Peng Cao / Yuansheng Liu / Ailing Yu / Dong Wang / Lei Chen / Rajamanikandan Sundarraj / Zhiguang Yuchi / Yong Gong / Hans Merzendorfer / Qing Yang /   Abstract: Chitin, the most abundant aminopolysaccharide in nature, is an extracellular polymer consisting of N-acetylglucosamine (GlcNAc) units. The key reactions of chitin biosynthesis are catalysed by chitin ...Chitin, the most abundant aminopolysaccharide in nature, is an extracellular polymer consisting of N-acetylglucosamine (GlcNAc) units. The key reactions of chitin biosynthesis are catalysed by chitin synthase, a membrane-integrated glycosyltransferase that transfers GlcNAc from UDP-GlcNAc to a growing chitin chain. However, the precise mechanism of this process has yet to be elucidated. Here we report five cryo-electron microscopy structures of a chitin synthase from the devastating soybean root rot pathogenic oomycete Phytophthora sojae (PsChs1). They represent the apo, GlcNAc-bound, nascent chitin oligomer-bound, UDP-bound (post-synthesis) and chitin synthase inhibitor nikkomycin Z-bound states of the enzyme, providing detailed views into the multiple steps of chitin biosynthesis and its competitive inhibition. The structures reveal the chitin synthesis reaction chamber that has the substrate-binding site, the catalytic centre and the entrance to the polymer-translocating channel that allows the product polymer to be discharged. This arrangement reflects consecutive key events in chitin biosynthesis from UDP-GlcNAc binding and polymer elongation to the release of the product. We identified a swinging loop within the chitin-translocating channel, which acts as a 'gate lock' that prevents the substrate from leaving while directing the product polymer into the translocating channel for discharge to the extracellular side of the cell membrane. This work reveals the directional multistep mechanism of chitin biosynthesis and provides a structural basis for inhibition of chitin synthesis. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32918.map.gz emd_32918.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32918-v30.xml emd-32918-v30.xml emd-32918.xml emd-32918.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32918.png emd_32918.png | 151.2 KB | ||
| Filedesc metadata |  emd-32918.cif.gz emd-32918.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32918 http://ftp.pdbj.org/pub/emdb/structures/EMD-32918 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32918 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32918 | HTTPS FTP |
-Validation report
| Summary document |  emd_32918_validation.pdf.gz emd_32918_validation.pdf.gz | 368.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32918_full_validation.pdf.gz emd_32918_full_validation.pdf.gz | 367.8 KB | Display | |
| Data in XML |  emd_32918_validation.xml.gz emd_32918_validation.xml.gz | 6.4 KB | Display | |
| Data in CIF |  emd_32918_validation.cif.gz emd_32918_validation.cif.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32918 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32918 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32918 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32918 | HTTPS FTP |
-Related structure data
| Related structure data |  7x06MC  7wjmC  7wjnC  7wjoC  7x05C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32918.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32918.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of chitin synthase 1 from Phytophthora sojae complexed with UDP | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
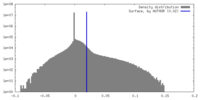
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Chitin synthase 1
| Entire | Name: Chitin synthase 1 |
|---|---|
| Components |
|
-Supramolecule #1: Chitin synthase 1
| Supramolecule | Name: Chitin synthase 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Phytophthora sojae strain P6497 (eukaryote) Phytophthora sojae strain P6497 (eukaryote) |
-Macromolecule #1: Chitin synthase
| Macromolecule | Name: Chitin synthase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: chitin synthase |
|---|---|
| Source (natural) | Organism:  Phytophthora sojae strain P6497 (eukaryote) / Strain: P6497 Phytophthora sojae strain P6497 (eukaryote) / Strain: P6497 |
| Molecular weight | Theoretical: 103.146703 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSGAPPPSSG FAPRSYGQQP LSHAPRSSMM SVEYDGIPLP PPSIRSCGSQ QYVTSYIPTG AAFPPSSVQD MISSMKSYAS ATDLVRTYS EIPSVEEALS TLDRAAAALN ARRYRDALKL YLEGGYAMAN VAERQANPKI CNLLTSKGFE TLNWCARLCD W IEGRIKEK ...String: MSGAPPPSSG FAPRSYGQQP LSHAPRSSMM SVEYDGIPLP PPSIRSCGSQ QYVTSYIPTG AAFPPSSVQD MISSMKSYAS ATDLVRTYS EIPSVEEALS TLDRAAAALN ARRYRDALKL YLEGGYAMAN VAERQANPKI CNLLTSKGFE TLNWCARLCD W IEGRIKEK HPRPGVHKVG IPVSNWDEDW VGPFMDEEEA RRMWYTPVYC PHPIDFSNLG YRLRCVETGR RPRLMICITM YN EGPQQLK ATLKKLANNL AYLKEQMPGD EKSLTGAFAG DDVWQNVLVC IVADGREQVH PKTLDYLEAI GLYDEDLLTI NSA GIGAQC HLFEHTLQLS VNGKCLLPIQ TVFALKENKA SKLDSHHWYF NAFAEQIQPE YTAVMDVGTM LTKSALYHLL FAFE RNHQI GGACGQLTVD NPFENLSNWV ISAQHFEYKI SNILDKSLES CFGFISVLPG AFSAYRYEAI RGAPLDAYFQ TLNIE LDVL GPFIGNMYLA EDRILSFEVV ARKNCNWTMH YVKDAVARTD VPHDLVGLIS QRKRWLNGAF FATLFSIWNW GRIYSE SKH TFVRKMAFLV FYVYHLLYTA FGFFLPANLY LALFFIVFQG FQQNRLEFID TSEYSQTVLD CAVYIYNFSY LFGLLML II IGLGNNPKHM KLTYYFVGAV FGLMMMLSSL VGAGIFFSTP ATVHSIVVSI LTVGVYFIAS ALHGEVHHIF MTFTHYTA L IPSFVNIFTI YSFCNLQDLS WGTKGLHDDP LLAASLDETE KGDFKDVIAK RRALEELRRE EKERVENRKK NFEAFRTNV LLTWAFSNLI FALFVVYFAS SSTYMPVLYI FVASLNTCRL LGSIGHWVYI HTEGLRGRVI DKSECGNGTG RYPQNSYVQL EEHYAALAE DQRTYASGRT NASVRTVNDV SSAA UniProtKB: Chitin synthase 1 |
-Macromolecule #2: URIDINE-5'-DIPHOSPHATE
| Macromolecule | Name: URIDINE-5'-DIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: UDP |
|---|---|
| Molecular weight | Theoretical: 404.161 Da |
| Chemical component information |  ChemComp-UDP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)