+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
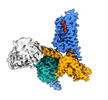
| Title | Structure of an orphan GPCR-G protein signaling complex | |||||||||
 Map data Map data | Structure of an orphan GPCR-G protein signaling complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPR88 / signaling complex / cryo-EM / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmotor learning / G protein-coupled photoreceptor activity / cellular response to light stimulus / beta2-adrenergic receptor activity / neuromuscular process controlling balance / ciliary membrane / cytoskeletal motor activity / phototransduction / neuronal action potential / adenylate cyclase inhibitor activity ...motor learning / G protein-coupled photoreceptor activity / cellular response to light stimulus / beta2-adrenergic receptor activity / neuromuscular process controlling balance / ciliary membrane / cytoskeletal motor activity / phototransduction / neuronal action potential / adenylate cyclase inhibitor activity / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / response to prostaglandin E / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / adenylate cyclase regulator activity / adenylate cyclase-inhibiting serotonin receptor signaling pathway / cellular response to forskolin / regulation of mitotic spindle organization / Regulation of insulin secretion / locomotory behavior / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / response to peptide hormone / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / centriolar satellite / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / GDP binding / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / Extra-nuclear estrogen signaling / cell population proliferation / cilium / ciliary basal body / G protein-coupled receptor signaling pathway / cell division / lysosomal membrane / GTPase activity / synapse / centrosome / GTP binding / protein-containing complex binding / nucleolus / magnesium ion binding / Golgi apparatus / signal transduction / extracellular exosome Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Xu J / Chen G | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Activation and allosteric regulation of the orphan GPR88-Gi1 signaling complex. Authors: Geng Chen / Jun Xu / Asuka Inoue / Maximilian F Schmidt / Chen Bai / Qiuyuan Lu / Peter Gmeiner / Zheng Liu / Yang Du /     Abstract: GPR88 is an orphan class A G-protein-coupled receptor that is highly expressed in the striatum and regulates diverse brain and behavioral functions. Here we present cryo-EM structures of the human ...GPR88 is an orphan class A G-protein-coupled receptor that is highly expressed in the striatum and regulates diverse brain and behavioral functions. Here we present cryo-EM structures of the human GPR88-Gi1 signaling complex with or without a synthetic agonist (1R, 2R)-2-PCCA. We show that (1R, 2R)-2-PCCA is an allosteric modulator binding to a herein identified pocket formed by the cytoplasmic ends of transmembrane segments 5, 6, and the extreme C terminus of the α5 helix of Gi1. We also identify an electron density in the extracellular orthosteric site that may represent a putative endogenous ligand of GPR88. These structures, together with mutagenesis studies and an inactive state model obtained from metadynamics simulations, reveal a unique activation mechanism for GPR88 with a set of distinctive structure features and a water-mediated polar network. Overall, our results provide a structural framework for understanding the ligand binding, activation and signaling mechanism of GPR88, and will facilitate the innovative drug discovery for neuropsychiatric disorders and for deorphanization of this receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32904.map.gz emd_32904.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32904-v30.xml emd-32904-v30.xml emd-32904.xml emd-32904.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32904.png emd_32904.png | 104.1 KB | ||
| Filedesc metadata |  emd-32904.cif.gz emd-32904.cif.gz | 6.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32904 http://ftp.pdbj.org/pub/emdb/structures/EMD-32904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32904 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32904 | HTTPS FTP |
-Related structure data
| Related structure data |  7wz4MC  7ejxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32904.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32904.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of an orphan GPCR-G protein signaling complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : GPR88-Gi1 signaling complex
| Entire | Name: GPR88-Gi1 signaling complex |
|---|---|
| Components |
|
-Supramolecule #1: GPR88-Gi1 signaling complex
| Supramolecule | Name: GPR88-Gi1 signaling complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: GPR88-Gi1
| Supramolecule | Name: GPR88-Gi1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: scFv16
| Supramolecule | Name: scFv16 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Probable G-protein coupled receptor 88
| Macromolecule | Name: Probable G-protein coupled receptor 88 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.697926 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDKLE VLFQGPGSAD LEDNWETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPPKLEDKS PDSPEMKDFR HGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLMTNSS STSTSSTTGG SLLLLCEEEE SWAGRRIPVS L LYSGLAIG ...String: DYKDDDDKLE VLFQGPGSAD LEDNWETLND NLKVIEKADN AAQVKDALTK MRAAALDAQK ATPPKLEDKS PDSPEMKDFR HGFDILVGQ IDDALKLANE GKVKEAQAAA EQLKTTRNAY IQKYLMTNSS STSTSSTTGG SLLLLCEEEE SWAGRRIPVS L LYSGLAIG GTLANGMVIY LVSSFRKLQT TSNAFIVNGC AADLSVCALW MPQEAVLGLL PTGSAEPPAD WDGAGGSYRL LR GGLLGLG LTVSLLSHCL VALNRYLLIT RAPATYQALY QRRHTAGMLA LSWALALGLV LLLPPWAPRP GAAPPRVHYP ALL AAAALL AQTALLLHCY LGIVRRVRVS VKRVSVLNFH LLHQLPGCAA AAAAFPGAQH APGPGGAAHP AQAQPLPPAL HPRR AQRRL SGLSVLLLCC VFLLATQPLV WVSLASGFSL PVPWGVQAAS WLLCCALSAL NPLLYTWRNE EFRRSVRSVL PGVGD AAAA AVAATAVPAV SQAQLGTRAA GQHWLEHHHH HHHHHH UniProtKB: G protein-coupled receptor 88 |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.415031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKSTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGGQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHESM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCA TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.286891 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHLEVL FQGPGSSGSE LDQLRQEAEQ LKNQIRDARK ACADATLSQI TNNIDPVGRI QMRTRRTLRG HLAKIYAMHW GTDSRLLVS ASQDGKLIIW DSYTTNKVHA IPLRSSWVMT CAYAPSGNYV ACGGLDNICS IYNLKTREGN VRVSRELAGH T GYLSCCRF ...String: HHHHHHLEVL FQGPGSSGSE LDQLRQEAEQ LKNQIRDARK ACADATLSQI TNNIDPVGRI QMRTRRTLRG HLAKIYAMHW GTDSRLLVS ASQDGKLIIW DSYTTNKVHA IPLRSSWVMT CAYAPSGNYV ACGGLDNICS IYNLKTREGN VRVSRELAGH T GYLSCCRF LDDNQIVTSS GDTTCALWDI ETGQQTTTFT GHTGDVMSLS LAPDTRLFVS GACDASAKLW DVREGMCRQT FT GHESDIN AICFFPNGNA FATGSDDATC RLFDLRADQE LMTYSHDNII CGITSVSFSK SGRLLLAGYD DFNCNVWDAL KAD RAGVLA GHDNRVSCLG VTDDGMAVAT GSWDSFLKIW N UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.784896 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KAAAHHHHHH HH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Sugar embedding | Material: vitreous ice |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.0 µm / Calibrated defocus min: 0.8 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.4000000000000001 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 314834 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















