+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Choline transporter-like protein 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | choline binding / cholesterol binding / transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationethanolamine transport / ethanolamine transmembrane transporter activity / Choline catabolism / : / choline catabolic process / choline transmembrane transporter activity / phosphatidylcholine biosynthetic process / choline transport / antiporter activity / Synthesis of PC ...ethanolamine transport / ethanolamine transmembrane transporter activity / Choline catabolism / : / choline catabolic process / choline transmembrane transporter activity / phosphatidylcholine biosynthetic process / choline transport / antiporter activity / Synthesis of PC / transport across blood-brain barrier / transmembrane transport / mitochondrial outer membrane / mitochondrion / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.86 Å | |||||||||
 Authors Authors | Chi XM / Zhou Q | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: Rational exploration of fold atlas for human solute carrier proteins. Authors: Tengyu Xie / Ximin Chi / Bangdong Huang / Fangfei Ye / Qiang Zhou / Jing Huang /  Abstract: The solute carrier (SLC) superfamily is the largest group of proteins responsible for the transmembrane transport of substances in human cells. It includes more than 400 members that are organized ...The solute carrier (SLC) superfamily is the largest group of proteins responsible for the transmembrane transport of substances in human cells. It includes more than 400 members that are organized into 65 families according to their physiological function and sequence similarity. Different families of SLCs can adopt the same or different folds that determine the mechanism and reflect the evolutionary relationship between SLC members. Analysis of structural data in the literature before this work showed 13 different folds in the SLC superfamily covering 40 families and 343 members. To further study their mechanism, we systematically explored the SLC superfamily to look for more folds. Based on our results, at least three new folds are found for the SLC superfamily, one of which is in the choline-like transporter family (SLC44) and has been experimentally verified. Our work has laid a foundation and provided important insights for the systematic and comprehensive study of the structure and function of SLC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32865.map.gz emd_32865.map.gz | 36.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32865-v30.xml emd-32865-v30.xml emd-32865.xml emd-32865.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32865_fsc.xml emd_32865_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_32865.png emd_32865.png | 80.8 KB | ||
| Filedesc metadata |  emd-32865.cif.gz emd-32865.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32865 http://ftp.pdbj.org/pub/emdb/structures/EMD-32865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32865 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32865 | HTTPS FTP |
-Related structure data
| Related structure data |  7wwbMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32865.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32865.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.087 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Choline transporter-like protein 1 in complex with choline
| Entire | Name: Choline transporter-like protein 1 in complex with choline |
|---|---|
| Components |
|
-Supramolecule #1: Choline transporter-like protein 1 in complex with choline
| Supramolecule | Name: Choline transporter-like protein 1 in complex with choline type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Choline transporter-like protein 1
| Macromolecule | Name: Choline transporter-like protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.369844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGCCSSASSA AQSSKREWKP LEDRSCTDIP WLLLFILFCI GMGFICGFSI ATGAAARLVS GYDSYGNICG QKNTKLEAIP NSGMDHTQR KYVFFLDPCN LDLINRKIKS VALCVAACPR QELKTLSDVQ KFAEINGSAL CSYNLKPSEY TTSPKSSVLC P KLPVPASA ...String: MGCCSSASSA AQSSKREWKP LEDRSCTDIP WLLLFILFCI GMGFICGFSI ATGAAARLVS GYDSYGNICG QKNTKLEAIP NSGMDHTQR KYVFFLDPCN LDLINRKIKS VALCVAACPR QELKTLSDVQ KFAEINGSAL CSYNLKPSEY TTSPKSSVLC P KLPVPASA PIPFFHRCAP VNISCYAKFA EALITFVSDN SVLHRLISGV MTSKEIILGL CLLSLVLSMI LMVIIRYISR VL VWILTIL VILGSLGGTG VLWWLYAKQR RSPKETVTPE QLQIAEDNLR ALLIYAISAT VFTVILFLIM LVMRKRVALT IAL FHVAGK VFIHLPLLVF QPFWTFFALV LFWVYWIMTL LFLGTTGSPV QNEQGFVEFK ISGPLQYMWW YHVVGLIWIS EFIL ACQQM TVAGAVVTYY FTRDKRNLPF TPILASVNRL IRYHLGTVAK GSFIITLVKI PRMILMYIHS QLKGKENACA RCVLK SCIC CLWCLEKCLN YLNQNAYTAT AINSTNFCTS AKDAFVILVE NALRVATINT VGDFMLFLGK VLIVCSTGLA GIMLLN YQQ DYTVWVLPLI IVCLFAFLVA HCFLSIYEMV VDVLFLCFAI DTKYNDGSPG REFYMDKVLM EFVENSRKAM KEAGKGG VA DSRELKPMAS GASSA UniProtKB: Choline transporter-like protein 1 |
-Macromolecule #2: CHOLINE ION
| Macromolecule | Name: CHOLINE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: CHT |
|---|---|
| Molecular weight | Theoretical: 104.171 Da |
| Chemical component information | 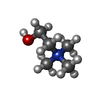 ChemComp-CHT: |
-Macromolecule #3: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 3 / Number of copies: 2 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.9000000000000001 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















