+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Gid12 bound GIDSR4 E3 ubiquitin ligase complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibited E3 ubiquitin Ligase / beta-propellor / LIGASE | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.8 Å | |||||||||
 Authors Authors | Qiao S / Cheng DJ / Schulman BA | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation. Authors: Shuai Qiao / Chia-Wei Lee / Dawafuti Sherpa / Jakub Chrustowicz / Jingdong Cheng / Maximilian Duennebacke / Barbara Steigenberger / Ozge Karayel / Duc Tung Vu / Susanne von Gronau / Matthias ...Authors: Shuai Qiao / Chia-Wei Lee / Dawafuti Sherpa / Jakub Chrustowicz / Jingdong Cheng / Maximilian Duennebacke / Barbara Steigenberger / Ozge Karayel / Duc Tung Vu / Susanne von Gronau / Matthias Mann / Florian Wilfling / Brenda A Schulman /    Abstract: Protein degradation, a major eukaryotic response to cellular signals, is subject to numerous layers of regulation. In yeast, the evolutionarily conserved GID E3 ligase mediates glucose-induced ...Protein degradation, a major eukaryotic response to cellular signals, is subject to numerous layers of regulation. In yeast, the evolutionarily conserved GID E3 ligase mediates glucose-induced degradation of fructose-1,6-bisphosphatase (Fbp1), malate dehydrogenase (Mdh2), and other gluconeogenic enzymes. "GID" is a collection of E3 ligase complexes; a core scaffold, RING-type catalytic core, and a supramolecular assembly module together with interchangeable substrate receptors select targets for ubiquitylation. However, knowledge of additional cellular factors directly regulating GID-type E3s remains rudimentary. Here, we structurally and biochemically characterize Gid12 as a modulator of the GID E3 ligase complex. Our collection of cryo-EM reconstructions shows that Gid12 forms an extensive interface sealing the substrate receptor Gid4 onto the scaffold, and remodeling the degron binding site. Gid12 also sterically blocks a recruited Fbp1 or Mdh2 from the ubiquitylation active sites. Our analysis of the role of Gid12 establishes principles that may more generally underlie E3 ligase regulation. #1:  Journal: Mol Cell / Year: 2021 Journal: Mol Cell / Year: 2021Title: GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme Authors: Sherpa D / Chrustowicz J / Qiao S / Langlois CR / Hehl LA / Gottemukkala KV / Hansen FM / Karayel O / von Gronau S / Prabu JR / Mann M / Alpi AF / Schulman BA | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32831.map.gz emd_32831.map.gz | 42.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32831-v30.xml emd-32831-v30.xml emd-32831.xml emd-32831.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32831.png emd_32831.png | 43.3 KB | ||
| Filedesc metadata |  emd-32831.cif.gz emd-32831.cif.gz | 7.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32831 http://ftp.pdbj.org/pub/emdb/structures/EMD-32831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32831 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32831 | HTTPS FTP |
-Validation report
| Summary document |  emd_32831_validation.pdf.gz emd_32831_validation.pdf.gz | 400.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32831_full_validation.pdf.gz emd_32831_full_validation.pdf.gz | 400.5 KB | Display | |
| Data in XML |  emd_32831_validation.xml.gz emd_32831_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  emd_32831_validation.cif.gz emd_32831_validation.cif.gz | 6.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32831 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32831 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32831 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32831 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32831.map.gz / Format: CCP4 / Size: 47.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32831.map.gz / Format: CCP4 / Size: 47.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.612 Å | ||||||||||||||||||||||||||||||||||||
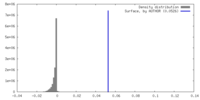
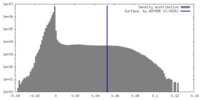
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Gid12-GIDSR4
| Entire | Name: Gid12-GIDSR4 |
|---|---|
| Components |
|
-Supramolecule #1: Gid12-GIDSR4
| Supramolecule | Name: Gid12-GIDSR4 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Gid1
| Macromolecule | Name: Gid1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MSEYMDDVDR EFINCLFPSY LLQQPVAYDL WILYLQHRKL FHKLKNTNLI NADENPTGVG MGRTKLTALT RKEIWSKLMN LGVLGTISFE AVNDDYLIQV YKYFYPDVND FTLRFGVKDS NKNSVRVMKA SSDMRKNAQE LLEPVLSERE MALNSNTSLE NDRNDDDDDD ...String: MSEYMDDVDR EFINCLFPSY LLQQPVAYDL WILYLQHRKL FHKLKNTNLI NADENPTGVG MGRTKLTALT RKEIWSKLMN LGVLGTISFE AVNDDYLIQV YKYFYPDVND FTLRFGVKDS NKNSVRVMKA SSDMRKNAQE LLEPVLSERE MALNSNTSLE NDRNDDDDDD DDDDDDDDDD DDDDDESDLE SLEGEVDTDT DDNNEGDGSD NHEEGGEEGS RGADADVSSA QQRAERVADP WIYQRSRSAI NIETESRNLW DTSDKNSGLQ YYPPDQSPSS SFSSPRVSSG NDKNDNEATN VLSNSGSKKK NSMIPDIYKI LGYFLPSRWQ AQPNNSLQLS QDGITHLQPN PDYHSYMTYE RSSASSASTR NRLRTSFENS GKVDFAVTWA NKSLPDNKLT IFYYEIKVLS VTSTESAENS NIVIGYKLVE NELMEATTKK SVSRSSVAGS SSSLGGSNNM SSNRVPSTSF TMEGTQRRDY IYEGGVSAMS LNVDGSINKC QKYGFDLNVF GYCGFDGLIT NSTEQSKEYA KPFGRDDVIG CGINFIDGSI FFTKNGIHLG NAFTDLNDLE FVPYVALRPG NSIKTNFGLN EDFVFDIIGY QDKWKSLAYE HICRGRQMDV SIEEFDSDES EEDETENGPE ENKSTNVNED LMDIDQEDGA AGNKDTKKLN DEKDNNLKFL LGEDNRFIDG KLVRPDVNNI NNLSVDDGSL PNTLNVMIND YLIHEGLVDV AKGFLKDLQK DAVNVNGQHS ESKDVIRHNE RQIMKEERMV KIRQELRYLI NKGQISKCIN YIDNEIPDLL KNNLELVFEL KLANYLVMIK KSSSKDDDEI ENLILKGQEL SNEFIYDTKI PQSLRDRFSG QLSNVSALLA YSNPLVEAPK EISGYLSDEY LQERLFQVSN NTILTFLHKD SECALENVIS NTRAMLSTLL EYNAFGSTNS SDPRYYKAIN FDEDVLNL |
-Macromolecule #2: Gid2
| Macromolecule | Name: Gid2 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MSELLDSFET EFAKFYTDSN LEETNLQKCL DHTHEFKSQL KKLKAHLNKH IQESKPEVYN KLSDKEKQKF KRKRELIIEK LSKSQRQWDH SVKKQIKYVS QQSNRFNKST LNKLKEFDID SVYVNKLPKE TMENVNEAIG YHILRYSIDN MPLGNKNEAF QYLKDVYGIT ...String: MSELLDSFET EFAKFYTDSN LEETNLQKCL DHTHEFKSQL KKLKAHLNKH IQESKPEVYN KLSDKEKQKF KRKRELIIEK LSKSQRQWDH SVKKQIKYVS QQSNRFNKST LNKLKEFDID SVYVNKLPKE TMENVNEAIG YHILRYSIDN MPLGNKNEAF QYLKDVYGIT NKESTEFIEM GQIVHDLKKG DTESCLKWCS NEMESLSSNH TALSSLKFDL YTLSAMQIVK HGNPVELYYQ ITQNAPLDCF RHREKELMQN VVPLLTKSLI GQPIEDIDSK VNKELKECTS LFIKEYCAAK HIFFDSPLFL IVLSGLISFQ FFIKYKTIRE LAHVDWTTKD ELPFDVKLPD FLTHFHPIFI CPVLKEETTT ENPPYSLACH HIISKKALDR LSKNGTITFK CPYCPVNTSM SSTKKVRFVM L |
-Macromolecule #3: Gid4
| Macromolecule | Name: Gid4 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MINNPKVDSV AEKPKAVTSK QSEQAASPEP TPAPPVSRNQ YPITFNLTST APFHLHDRHR YLQEQDLYKC ASRDSLSSLQ QLAHTPNGST RKKYIVEDQS PYSSENPVIV TSSYNHTVCT NYLRPRMQFT GYQISGYKRY QVTVNLKTVD LPKKDCTSLS PHLSGFLSIR ...String: MINNPKVDSV AEKPKAVTSK QSEQAASPEP TPAPPVSRNQ YPITFNLTST APFHLHDRHR YLQEQDLYKC ASRDSLSSLQ QLAHTPNGST RKKYIVEDQS PYSSENPVIV TSSYNHTVCT NYLRPRMQFT GYQISGYKRY QVTVNLKTVD LPKKDCTSLS PHLSGFLSIR GLTNQHPEIS TYFEAYAVNH KELGFLSSSW KDEPVLNEFK ATDQTDLEHW INFPSFRQLF LMSQKNGLNS TDDNGTTNAA KKLPPQQLPT TPSADAGNIS RIFSQEKQFD NYLNERFIFM KWKEKFLVPD ALLMEGVDGA SYDGFYYIVH DQVTGNIQGF YYHQDAEKFQ QLELVPSLKN KVESSDCSFE FA |
-Macromolecule #4: Gid5
| Macromolecule | Name: Gid5 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MTVAYSLENL KKISNSLVGD QLAKVDYFLA PKCQIFQCLL SIEQSDGVEL KNAKLDLLYT LLHLEPQQRD IVGTYYFDIV SAIYKSMSLA SSFTKNNSST NYKYIKLLNL CAGVYPNCGF PDLQYLQNGF IQLVNHKFLR SKCKIDEVVT IIELLKLFLL VDEKNCSDFN ...String: MTVAYSLENL KKISNSLVGD QLAKVDYFLA PKCQIFQCLL SIEQSDGVEL KNAKLDLLYT LLHLEPQQRD IVGTYYFDIV SAIYKSMSLA SSFTKNNSST NYKYIKLLNL CAGVYPNCGF PDLQYLQNGF IQLVNHKFLR SKCKIDEVVT IIELLKLFLL VDEKNCSDFN KSKFMEEERE VTETSHYQDF KMAESLEHII VKISSKYLDQ ISLKYIVRLK VSRPASPSSV KNDPFDNKGV DCTRAIPKKI NISNMYDSSL LSLALLLYLR YHYMIPGDRK LRNDATFKMF VLGLLKSNDV NIRCVALKFL LQPYFTEDKK WEDTRTLEKI LPYLVKSFNY DPLPWWFDPF DMLDSLIVLY NEITPMNNPV LTTLAHTNVI FCILSRFAQC LSLPQHNEAT LKTTTKFIKI CASFAASDEK YRLLLLNDTL LLNHLEYGLE SHITLIQDFI SLKDEIKETT TESHSMCLPP IYDHDFVAAW LLLLKSFSRS VSALRTTLKR NKIAQLLLQI LSKTYTLTKE CYFAGQDFMK PEIMIMGITL GSICNFVVEF SNLQSFMLRN GIIDIIEKML TDPLFNSKKA WDDNEDERRI ALQGIPVHEV KANSLWVLRH LMYNCQNEEK FQLLAKIPMN LILDFINDPC WAVQAQCFQL LRNLTCNSRK IVNILLEKFK DVEYKIDPQT GNKISIGSTY LFEFLAKKMR LLNPLDTQQK KAMEGILYII VNLAAVNENK KQLVIEQDEI LNIMSEILVE TTTDSSSYGN DSNLKLACLW VLNNLLWNSS VSHYTQYAIE NGLEPGHSPS DSENPQSTVT IGYNESVAGG YSRGKYYDEP DGDDSSSNAN DDEDDDNDEG DDEGDEFVRT PAAKGSTSNV QVTRATVERC RKLVEVGLYD LVRKNITDES LSVREKARTL LYHMDLLLKV K |
-Macromolecule #5: Gid8
| Macromolecule | Name: Gid8 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MTISTLSNET TKSGSCSGQG KNGGKDFTYG KKCFTKEEWK EQVAKYSAMG ELYANKTIHY PLKIQPNSSG GSQDEGFATI QTTPIEPTLP RLLLNYFVSM AYEDSSIRMA KELGFIRNNK DIAVFNDLYK IKERFHIKHL IKLGRINEAM EEINSIFGLE VLEETFNATG ...String: MTISTLSNET TKSGSCSGQG KNGGKDFTYG KKCFTKEEWK EQVAKYSAMG ELYANKTIHY PLKIQPNSSG GSQDEGFATI QTTPIEPTLP RLLLNYFVSM AYEDSSIRMA KELGFIRNNK DIAVFNDLYK IKERFHIKHL IKLGRINEAM EEINSIFGLE VLEETFNATG SYTGRTDRQQ QQQQQQFDID GDLHFKLLLL NLIEMIRSHH QQENITKDSN DFILNLIQYS QNKLAIKASS SVKKMQELEL AMTLLLFPLS DSADSGSIKL PKSLQNLYSI SLRSKIADLV NEKLLKFIHP RIQFEISNNN SKFPDLLNSD KKIITQNFTV YNNNLVNGSN GTKITHISSD QPINEKMSSN EVTAAANSVW LNQRDGNVGT GSAATTFHNL ENKNYWNQTS ELLSSSNGKE KGLEFNNYYS SEFPYEPRLT QIMKLWCWCE NQLHHNQIGV PRVEN |
-Macromolecule #6: Gid9
| Macromolecule | Name: Gid9 / type: protein_or_peptide / ID: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MAEKSIFNEP DVDFHLKLNQ QLFHIPYELL SKRIKHTQAV INKETKSLHE HTAALNQIFE HNDVEHDELA LAKITEMIRK VDHIERFLNT QIKSYCQILN RIKKRLEFFH ELKDIKSQNS GTSHNGNNEG TRTKLIQWYQ SYTNILIGDY LTRNNPIKYN SETKDHWNSG ...String: MAEKSIFNEP DVDFHLKLNQ QLFHIPYELL SKRIKHTQAV INKETKSLHE HTAALNQIFE HNDVEHDELA LAKITEMIRK VDHIERFLNT QIKSYCQILN RIKKRLEFFH ELKDIKSQNS GTSHNGNNEG TRTKLIQWYQ SYTNILIGDY LTRNNPIKYN SETKDHWNSG VVFLKQSQLD DLIDYDVLLE ANRISTSLLH ERNLLPLISW INENKKTLTK KSSILEFQAR LQEYIELLKV DNYTDAIVCF QRFLLPFVKS NFTDLKLASG LLIFIKYCND QKPTSSTSSG FDTEEIKSQS LPMKKDRIFQ HFFHKSLPRI TSKPAVNTTD YDKSSLINLQ SGDFERYLNL LDDQRWSVLN DLFLSDFYSM YGISQNDPLL IYLSLGISSL KTRDCLHPSD DENGNQETET ATTAEKEVED LQLFTLHSLK RKNCPVCSET FKPITQALPF AHHIQSQLFE NPILLPNGNV YDSKKLKKLA KTLKKQNLIS LNPGQIMDPV DMKIFCESDS IKMYPT |
-Macromolecule #7: Gid12
| Macromolecule | Name: Gid12 / type: protein_or_peptide / ID: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) Spodoptera aff. frugiperda 1 BOLD-2017 (butterflies/moths) |
| Sequence | String: MATGRIQFAV STPCNTKGKP SGYRLFEFKN DRLALVPSER GCTKVDVNAN IQAFCYLRPN GRDTSISPDA THILDSCDYM VLAKSNGFIE IISNYQYKIK NGLRLAPSYI LRCTPEDFES NFFSDYMIAG LEYSQGLLYC CMCSGRIYVF VMNLPTDYIQ YKNMYNPMFP ...String: MATGRIQFAV STPCNTKGKP SGYRLFEFKN DRLALVPSER GCTKVDVNAN IQAFCYLRPN GRDTSISPDA THILDSCDYM VLAKSNGFIE IISNYQYKIK NGLRLAPSYI LRCTPEDFES NFFSDYMIAG LEYSQGLLYC CMCSGRIYVF VMNLPTDYIQ YKNMYNPMFP DCFFKVHHDN NTTHSSEEEK LFEGSTRYTG RSCSKHICYF LLPIEPSHLR SSPVVSSFCN MYQGLPIYRP SMYLHIERGI STFHINPLDR FCFMTVSPRS PLFIRKIILP LTYVTFLSTF ISLKNSIQGD TCGEILSWDN VAQQNGFGSL FSWISNKFTF DTDIINSTIW DDIVKYSGTG MLDSGIVWKQ RQGHAKDDIY ELFHTQDMLG SSRRNSSFST ASSEPRPLSR RRRESFQALT RDAFRERMDV PCSTKWELDS FIRGLRRNTF MVDFEIVEKI SHRNGNDGVN EDDNTTDESD ETMTSFLTDN YKKMDIVCID HFVTLSAFRP RYYDEPIIKI DSLSNKNGSE NGTNEEEWAE SQMKVDGQVI DDETAQFKQA LGNLCSFKKL FMLDDSLCFI LDTHGVLLIN RFEIKNTKNL LRNSKDTIRI IPHDFGLIND TIVIINDIDV GTDNVCALTF HLVVTSMAGE ITVLKGEFFK NCRLGRIKLC DSLKLNRKDR FVDKLALIDY DGLNAQKRRL DYDEKDLYTF IVKKVKRD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 6.5 / Details: 25mM MES pH6.5 + 500mM NaCl + 1mM DTT |
| Grid | Model: C-flat-1.2/1.3 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 39.0 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 1.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)