[English] 日本語
 Yorodumi
Yorodumi- EMDB-32664: Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nuclear pore complex / Saccharomyces cerevisiae / cryo-EM / TRANSPORT PROTEIN | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 12.03 Å | |||||||||
 Authors Authors | Li ZQ / Chen SJB / Zhao L / Sui SF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2022 Journal: Cell Res / Year: 2022Title: Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex. Authors: Zongqiang Li / Shuaijiabin Chen / Liang Zhao / Guoqiang Huang / Xiong Pi / Shan Sun / Peiyi Wang / Sen-Fang Sui /  Abstract: Nuclear pore complexes (NPCs) mediate bidirectional nucleocytoplasmic transport of substances in eukaryotic cells. However, the accurate molecular arrangement of NPCs remains enigmatic owing to their ...Nuclear pore complexes (NPCs) mediate bidirectional nucleocytoplasmic transport of substances in eukaryotic cells. However, the accurate molecular arrangement of NPCs remains enigmatic owing to their huge size and highly dynamic nature. Here we determined the structure of the asymmetric unit of the inner ring (IR monomer) at 3.73 Å resolution by single-particle cryo-electron microscopy, and created an atomic model of the intact IR consisting of 192 molecules of 8 nucleoporins. In each IR monomer, the Z-shaped Nup188-Nup192 complex in the middle layer is sandwiched by two approximately parallel rhomboidal structures in the inner and outer layers, while Nup188, Nup192 and Nic96 link all subunits to constitute a relatively stable IR monomer. In contrast, the intact IR is assembled by loose and instable interactions between IR monomers. These structures, together with previously reported structural information of IR, reveal two distinct interaction modes between IR monomers and extensive flexible connections in IR assembly, providing a structural basis for the stability and malleability of IR. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32664.map.gz emd_32664.map.gz | 62.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32664-v30.xml emd-32664-v30.xml emd-32664.xml emd-32664.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32664.png emd_32664.png | 53.7 KB | ||
| Filedesc metadata |  emd-32664.cif.gz emd-32664.cif.gz | 3.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32664 http://ftp.pdbj.org/pub/emdb/structures/EMD-32664 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32664 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32664 | HTTPS FTP |
-Validation report
| Summary document |  emd_32664_validation.pdf.gz emd_32664_validation.pdf.gz | 410.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32664_full_validation.pdf.gz emd_32664_full_validation.pdf.gz | 409.7 KB | Display | |
| Data in XML |  emd_32664_validation.xml.gz emd_32664_validation.xml.gz | 8.1 KB | Display | |
| Data in CIF |  emd_32664_validation.cif.gz emd_32664_validation.cif.gz | 9.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32664 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32664 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32664 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32664 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32664.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32664.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.672 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex
| Entire | Name: Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex
| Supramolecule | Name: Cryo-EM map of the whole Saccharomyces cerevisiae nuclear pore complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 12.03 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 51220 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


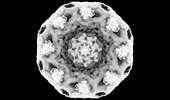










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















