[English] 日本語
 Yorodumi
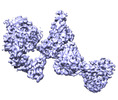
Yorodumi- EMDB-32641: Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-15 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-15 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-COV-2 / Spike / Antibody / Viral protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Zhan WQ / Zhang X / Chen ZG / Sun L | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2022 Journal: J Virol / Year: 2022Title: Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer. Authors: Wuqiang Zhan / Xiaolong Tian / Xiang Zhang / Shenghui Xing / Wenping Song / Qianying Liu / Aihua Hao / Yuxia Hu / Meng Zhang / Tianlei Ying / Zhenguo Chen / Fei Lan / Lei Sun /  Abstract: The continuous emergence of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants poses new challenges in the fight against the coronavirus disease 2019 (COVID-19) pandemic. The ...The continuous emergence of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants poses new challenges in the fight against the coronavirus disease 2019 (COVID-19) pandemic. The newly emerging Omicron strain caused serious immune escape and raised unprecedented concern all over the world. The development of an antibody targeting a conserved and universal epitope is urgently needed. A subset of neutralizing antibodies (NAbs) against COVID-19 from convalescent patients were isolated in our previous study. In this study, we investigated the accommodation of these NAbs to SARS-CoV-2 variants of concern (VOCs), revealing that IgG 553-49 neutralizes pseudovirus of the SARS-CoV-2 Omicron variant. In addition, we determined the cryo-electron microscopy (cryo-EM) structure of the SARS-CoV-2 spike (S) protein complexed with three monoclonal antibodies targeting different epitopes, including 553-49, 553-15, and 553-60. Notably, 553-49 targets a novel conserved epitope and neutralizes the virus by disassembling S trimers. IgG 553-15, an antibody that neutralizes all of the VOCs except Omicron, cross-links two S trimers to form a trimer dimer, demonstrating that 553-15 neutralizes the virus by steric hindrance and virion aggregation. These findings suggest the potential to develop 553-49 and other antibodies targeting this highly conserved epitope as promising therapeutic reagents for COVID-19. The emergence of the Omicron strain of SARS-CoV-2 caused higher immune escape, raising unprecedented concerns about the effectiveness of antibody therapies and vaccines. In this study, we identified a SARS-CoV-2 neutralizing antibody, 553-49, which neutralizes all variants by targeting a completely conserved novel epitope. In addition, we revealed that IgG 553-15 neutralizes SARS-CoV-2 by cross-linking virions and that 553-60 functions by blocking receptor binding. Comparison of different receptor binding domain (RBD) epitopes revealed that the 553-49 epitope is hidden in the S trimer and keeps a high degree of conservation during SARS-CoV-2 evolution, making 553-49 a promising therapeutic reagent against the emerging Omicron and future variants of SARS-CoV-2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32641.map.gz emd_32641.map.gz | 11.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32641-v30.xml emd-32641-v30.xml emd-32641.xml emd-32641.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32641.png emd_32641.png | 119.2 KB | ||
| Filedesc metadata |  emd-32641.cif.gz emd-32641.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32641 http://ftp.pdbj.org/pub/emdb/structures/EMD-32641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32641 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32641 | HTTPS FTP |
-Related structure data
| Related structure data |  7wo7MC  7wo4C  7wo5C  7woaC  7wobC  7wocC  7wogC  7wz1C  7wz2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32641.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32641.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Spike with mAb15
| Entire | Name: Spike with mAb15 |
|---|---|
| Components |
|
-Supramolecule #1: Spike with mAb15
| Supramolecule | Name: Spike with mAb15 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: mAb15 VH
| Macromolecule | Name: mAb15 VH / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.194086 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVQSGGG LVQPGGSLRL SCAASGFTFS SYWMSWVRQA PGKGLEWVAN INQDGGEKYY VDSVKGRFTI SRDNAKNSLF LQMNSVRAE DTAVYFCARV WYYYGPRDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG ...String: EVQLVQSGGG LVQPGGSLRL SCAASGFTFS SYWMSWVRQA PGKGLEWVAN INQDGGEKYY VDSVKGRFTI SRDNAKNSLF LQMNSVRAE DTAVYFCARV WYYYGPRDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKRVE PKSCDKT |
-Macromolecule #2: mAb15 VL
| Macromolecule | Name: mAb15 VL / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.315426 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQPHSV SESPGKTVTI SCTRSSGSIA SNYVQWYQQR PGSSPTTVIY EDNQRPSGVP DRFSGSIDSS SNSASLTISG LKTEDEADY YCQSYDGSNH NVVFGGGTEL TVLSQPKAAP SVTLFPPSSE ELQANKATLV CLISDFYPGA VTVAWKADSS P VKAGVETT ...String: DIVMTQPHSV SESPGKTVTI SCTRSSGSIA SNYVQWYQQR PGSSPTTVIY EDNQRPSGVP DRFSGSIDSS SNSASLTISG LKTEDEADY YCQSYDGSNH NVVFGGGTEL TVLSQPKAAP SVTLFPPSSE ELQANKATLV CLISDFYPGA VTVAWKADSS P VKAGVETT TPSKQSNNKY AASSYLSLTP EQWKSHRSYS CQVTHEGSTV EKTVAPTECS |
-Macromolecule #3: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.772391 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG VEGFNCYFPL Q SYGFQPTN ...String: NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAPG QTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG VEGFNCYFPL Q SYGFQPTN GVGYQPYRVV VLSFELLHAP ATVCGP UniProtKB: Spike glycoprotein |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 61.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




















