+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trichodesmium erythraeum cyanophycin synthetase 1 (TeCphA1) | |||||||||
 Map data Map data | Map without b-factor sharpening | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cyanophycin / Non-ribosomal peptide synthesis / ATP / Aspartate / Arginine / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationcyanophycin synthase (L-aspartate-adding) / cyanophycin synthase (L-arginine-adding) / cyanophycin synthetase activity (L-aspartate-adding) / cyanophycin synthetase activity (L-arginine-adding) / tetrahydrofolylpolyglutamate synthase activity / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Trichodesmium erythraeum IMS101 (bacteria) Trichodesmium erythraeum IMS101 (bacteria) | |||||||||
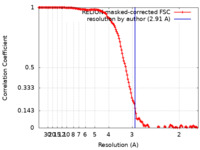
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Kawasaki M / Miyakawa T | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural bases for aspartate recognition and polymerization efficiency of cyanobacterial cyanophycin synthetase. Authors: Takuya Miyakawa / Jian Yang / Masato Kawasaki / Naruhiko Adachi / Ayumu Fujii / Yumiko Miyauchi / Tomonari Muramatsu / Toshio Moriya / Toshiya Senda / Masaru Tanokura /   Abstract: Cyanophycin is a natural biopolymer consisting of equimolar amounts of aspartate and arginine as the backbone and branched sidechain, respectively. It is produced by a single enzyme, cyanophycin ...Cyanophycin is a natural biopolymer consisting of equimolar amounts of aspartate and arginine as the backbone and branched sidechain, respectively. It is produced by a single enzyme, cyanophycin synthetase (CphA1), and accumulates as a nitrogen reservoir during N fixation by most cyanobacteria. A recent structural study showed that three constituent domains of CphA1 function as two distinct catalytic sites and an oligomerization interface in cyanophycin synthesis. However, it remains unclear how the ATP-dependent addition of aspartate to cyanophycin is initiated at the catalytic site of the glutathione synthetase-like domain. Here, we report the cryogenic electron microscopy structures of CphA1, including a complex with aspartate, cyanophycin primer peptide, and ATP analog. These structures reveal the aspartate binding mode and phosphate-binding loop movement to the active site required for the reaction. Furthermore, structural and mutational data show a potential role of protein dynamics in the catalytic efficiency of the arginine condensation reaction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32381.map.gz emd_32381.map.gz | 27.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32381-v30.xml emd-32381-v30.xml emd-32381.xml emd-32381.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32381_fsc.xml emd_32381_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_32381.png emd_32381.png | 176.3 KB | ||
| Masks |  emd_32381_msk_1.map emd_32381_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32381.cif.gz emd-32381.cif.gz | 6.7 KB | ||
| Others |  emd_32381_additional_1.map.gz emd_32381_additional_1.map.gz emd_32381_half_map_1.map.gz emd_32381_half_map_1.map.gz emd_32381_half_map_2.map.gz emd_32381_half_map_2.map.gz | 28.4 MB 27.1 MB 27.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32381 http://ftp.pdbj.org/pub/emdb/structures/EMD-32381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32381 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32381 | HTTPS FTP |
-Validation report
| Summary document |  emd_32381_validation.pdf.gz emd_32381_validation.pdf.gz | 885.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32381_full_validation.pdf.gz emd_32381_full_validation.pdf.gz | 885.2 KB | Display | |
| Data in XML |  emd_32381_validation.xml.gz emd_32381_validation.xml.gz | 16.9 KB | Display | |
| Data in CIF |  emd_32381_validation.cif.gz emd_32381_validation.cif.gz | 22.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32381 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32381 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32381 | HTTPS FTP |
-Related structure data
| Related structure data |  7wacMC  7wadC  7waeC  7wafC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32381.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32381.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map without b-factor sharpening | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_32381_msk_1.map emd_32381_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map with b-factor sharpening
| File | emd_32381_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map with b-factor sharpening | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_32381_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32381_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TeCphA1
| Entire | Name: TeCphA1 |
|---|---|
| Components |
|
-Supramolecule #1: TeCphA1
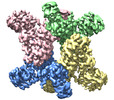
| Supramolecule | Name: TeCphA1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Homotetramer |
|---|---|
| Source (natural) | Organism:  Trichodesmium erythraeum IMS101 (bacteria) Trichodesmium erythraeum IMS101 (bacteria) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: Cyanophycin synthase
| Macromolecule | Name: Cyanophycin synthase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: cyanophycin synthase (L-aspartate-adding) |
|---|---|
| Source (natural) | Organism:  Trichodesmium erythraeum IMS101 (bacteria) / Strain: IMS101 Trichodesmium erythraeum IMS101 (bacteria) / Strain: IMS101 |
| Molecular weight | Theoretical: 99.079914 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKILKLQTLR GPNYWSIHRH KLVVMRLDLE DLYEKYTSDI PGFYKGLTEV LPSLVEHLCS PGVKGGFLTR VEKGTLIGHV IEHVAIELQ ELAGMPVGFG RTRETSTTGV FQVVIEYENE QAGRYAARAA VRLCQSIVDT GTYPATELQQ DLEDLKELKN Q ASLGPSTE ...String: MKILKLQTLR GPNYWSIHRH KLVVMRLDLE DLYEKYTSDI PGFYKGLTEV LPSLVEHLCS PGVKGGFLTR VEKGTLIGHV IEHVAIELQ ELAGMPVGFG RTRETSTTGV FQVVIEYENE QAGRYAARAA VRLCQSIVDT GTYPATELQQ DLEDLKELKN Q ASLGPSTE AIVKEAEARG IPWTQLGARF MIQFGYGVNQ KKIQATLSNQ TGILGVELAC DKEGTKRILK DAGVPVPRGT VA RYFDELQ DAIEYVGGYP IVIKPLDGNH GRGITIDVKN WQEAEEAYDL ARKASKTKTV IVERYYTGKD HRVLVVNGKV VAV AERVPA HVVGNGKSTI AELIEETNRD PQRGDGHDNI LTRITVDKSA LDILGKQGYS IDSIPLKGKK CFLRATANLS TGGI AVDRT DEIHPENVWL LSRVAKIIGL DIAGIDVVTE DISQPLREVE GVIVEVNAAP GFRMHVAPSR GLARNVAGAV MDMLF PGSK NGRIPILSVT GTNGKTTTTR LLAHIIKQTG KVVGYTTTDG TYIGEYLAET GDNTGPQSAH LILSDPTVEV AVLETA RGG ILRSGLGFSS CEVGIVLNVT ADHLGIGDID TIEQLAKLKS VVAESVMPKG YAVLNAEDPL VAAMADRVKG QVAYFSM DP NNELLLRHTE AGGLAAIYEN GYISILKGDW TLRIEKAVNV PITMAGKAPF MIANALAACL AVFTQGVKIE HIRKGLST F VASVDQTPGR MNMFNMGSYH ALVDYAHNPA SYEALGGFVR NWPGKRIGVV GGPGDRRDED FVSLGELAAD IFDEIIIKE DDDTRGRPRG NAAELICQGV KQFLNGIKNS ESKATYESIL DETAAINTAL DRAPIDGLVV ILPESVNRAI SLIEGRHVIQ DIELLQDSQ REPKDQEVLK SSILEHHHHH H UniProtKB: Cyanophycin synthetase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.19 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: The grid was washed by acetone prior to use. | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time was 15 seconds (blot force 0).. | ||||||||
| Details | This sample was mono-disperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2109 / Average exposure time: 55.75 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 120000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)