+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of nucleosome in complex with p300 acetyltransferase catalytic core (complex I) | |||||||||||||||||||||||||||||||||||||||
 マップデータ マップデータ | The cryo-EM map of p300-NCP complex I | |||||||||||||||||||||||||||||||||||||||
 試料 試料 |
| |||||||||||||||||||||||||||||||||||||||
 キーワード キーワード | p300 / nucleosome / acetyltransferase / GENE REGULATION | |||||||||||||||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報behavioral defense response / negative regulation of protein oligomerization / peptidyl-lysine propionylation / histone lactyltransferase (CoA-dependent) activity / peptidyl-lysine crotonylation / peptidyl-lysine butyrylation / histone butyryltransferase activity / swimming / histone H3K122 acetyltransferase activity / peptide butyryltransferase activity ...behavioral defense response / negative regulation of protein oligomerization / peptidyl-lysine propionylation / histone lactyltransferase (CoA-dependent) activity / peptidyl-lysine crotonylation / peptidyl-lysine butyrylation / histone butyryltransferase activity / swimming / histone H3K122 acetyltransferase activity / peptide butyryltransferase activity / regulation of tubulin deacetylation / histone H2B acetyltransferase activity / internal protein amino acid acetylation / peptide 2-hydroxyisobutyryltransferase activity / histone crotonyltransferase activity / protein propionyltransferase activity / NOTCH2 intracellular domain regulates transcription / thigmotaxis / L-lysine N-acetyltransferase activity, acting on acetyl phosphate as donor / positive regulation of TORC2 signaling / internal peptidyl-lysine acetylation / histone H4 acetyltransferase activity / cellular response to L-leucine / histone H3 acetyltransferase activity / NFE2L2 regulating ER-stress associated genes / acetylation-dependent protein binding / Activation of the TFAP2 (AP-2) family of transcription factors / NFE2L2 regulating inflammation associated genes / NGF-stimulated transcription / histone H3K18 acetyltransferase activity / N-terminal peptidyl-lysine acetylation / histone H3K27 acetyltransferase activity / LRR FLII-interacting protein 1 (LRRFIP1) activates type I IFN production / NFE2L2 regulates pentose phosphate pathway genes / STAT3 nuclear events downstream of ALK signaling / NFE2L2 regulating MDR associated enzymes / Polo-like kinase mediated events / host-mediated activation of viral transcription / TGFBR3 expression / regulation of androgen receptor signaling pathway / regulation of mitochondrion organization / Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells / Regulation of FOXO transcriptional activity by acetylation / RUNX3 regulates NOTCH signaling / NOTCH4 Intracellular Domain Regulates Transcription / Nuclear events mediated by NFE2L2 / Regulation of NFE2L2 gene expression / face morphogenesis / Regulation of gene expression by Hypoxia-inducible Factor / platelet formation / regulation of glycolytic process / NOTCH3 Intracellular Domain Regulates Transcription / TRAF6 mediated IRF7 activation / NFE2L2 regulating tumorigenic genes / NFE2L2 regulating anti-oxidant/detoxification enzymes / megakaryocyte development / protein-lysine-acetyltransferase activity / nuclear androgen receptor binding / protein acetylation / acyltransferase activity / STAT family protein binding / Formation of paraxial mesoderm / positive regulation of transforming growth factor beta receptor signaling pathway / acetyltransferase activity / FOXO-mediated transcription of cell death genes / fat cell differentiation / stimulatory C-type lectin receptor signaling pathway / RUNX1 interacts with co-factors whose precise effect on RUNX1 targets is not known / Zygotic genome activation (ZGA) / PI5P Regulates TP53 Acetylation / histone acetyltransferase complex / RUNX3 regulates p14-ARF / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / canonical NF-kappaB signal transduction / negative regulation of tumor necrosis factor-mediated signaling pathway / negative regulation of gluconeogenesis / NF-kappaB binding / pre-mRNA intronic binding / Attenuation phase / somitogenesis / positive regulation of T-helper 17 cell lineage commitment / positive regulation of DNA-binding transcription factor activity / histone acetyltransferase activity / cellular response to nutrient levels / negative regulation of protein-containing complex assembly / skeletal muscle tissue development / negative regulation of megakaryocyte differentiation / histone acetyltransferase / regulation of cellular response to heat / protein localization to CENP-A containing chromatin / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / Chromatin modifying enzymes / SARS-CoV-1 targets host intracellular signalling and regulatory pathways / Replacement of protamines by nucleosomes in the male pronucleus / Regulation of TP53 Activity through Acetylation / CENP-A containing nucleosome / : / Packaging Of Telomere Ends / Recognition and association of DNA glycosylase with site containing an affected purine / Cleavage of the damaged purine 類似検索 - 分子機能 | |||||||||||||||||||||||||||||||||||||||
| 生物種 |  Homo sapiens (ヒト) / synthetic construct (人工物) Homo sapiens (ヒト) / synthetic construct (人工物) | |||||||||||||||||||||||||||||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.95 Å | |||||||||||||||||||||||||||||||||||||||
 データ登録者 データ登録者 | Hatazawa S / Liu J / Takizawa Y / Zandian M / Negishi L / Kutateladze TG / Kurumizaka H | |||||||||||||||||||||||||||||||||||||||
| 資金援助 |  日本, 日本,  米国, 12件 米国, 12件
| |||||||||||||||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: iScience / 年: 2022 ジャーナル: iScience / 年: 2022タイトル: Structural basis for binding diversity of acetyltransferase p300 to the nucleosome. 著者: Suguru Hatazawa / Jiuyang Liu / Yoshimasa Takizawa / Mohamad Zandian / Lumi Negishi / Tatiana G Kutateladze / Hitoshi Kurumizaka /   要旨: p300 is a human acetyltransferase that associates with chromatin and mediates vital cellular processes. We now report the cryo-electron microscopy structures of the p300 catalytic core in complex ...p300 is a human acetyltransferase that associates with chromatin and mediates vital cellular processes. We now report the cryo-electron microscopy structures of the p300 catalytic core in complex with the nucleosome core particle (NCP). In the most resolved structure, the HAT domain and bromodomain of p300 contact nucleosomal DNA at superhelical locations 2 and 3, and the catalytic site of the HAT domain are positioned near the N-terminal tail of histone H4. Mutations of the p300-DNA interfacial residues of p300 substantially decrease binding to NCP. Three additional classes of p300-NCP complexes show different modes of the p300-NCP complex formation. Our data provide structural details critical to our understanding of the mechanism by which p300 acetylates multiple sites on the nucleosome. | |||||||||||||||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_32373.map.gz emd_32373.map.gz | 5.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-32373-v30.xml emd-32373-v30.xml emd-32373.xml emd-32373.xml | 19.5 KB 19.5 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
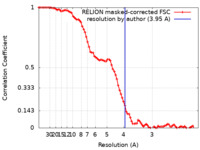
| FSC (解像度算出) |  emd_32373_fsc.xml emd_32373_fsc.xml | 8.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_32373.png emd_32373.png | 83.2 KB | ||
| Filedesc metadata |  emd-32373.cif.gz emd-32373.cif.gz | 7 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32373 http://ftp.pdbj.org/pub/emdb/structures/EMD-32373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32373 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32373 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_32373.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_32373.map.gz / 形式: CCP4 / 大きさ: 52.7 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | The cryo-EM map of p300-NCP complex I | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Nucleosome in complex with p300 acetyltransferase catalytic core ...
| 全体 | 名称: Nucleosome in complex with p300 acetyltransferase catalytic core (complex I) |
|---|---|
| 要素 |
|
-超分子 #1: Nucleosome in complex with p300 acetyltransferase catalytic core ...
| 超分子 | 名称: Nucleosome in complex with p300 acetyltransferase catalytic core (complex I) タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#4 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Histone H3.1
| 分子 | 名称: Histone H3.1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 15.305969 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEACEAY LVGLFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA UniProtKB: Histone H3.1 |
-分子 #2: Histone H4
| 分子 | 名称: Histone H4 / タイプ: protein_or_peptide / ID: 2 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 11.676703 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMSGRGKG GKGLGKGGAK RHRKVLRDNI QGITKPAIRR LARRGGVKRI SGLIYEETRG VLKVFLENVI RDAVTYTEHA KRKTVTAMD VVYALKRQGR TLYGFGG UniProtKB: Histone H4 |
-分子 #3: Histone H2A type 1-B/E
| 分子 | 名称: Histone H2A type 1-B/E / タイプ: protein_or_peptide / ID: 3 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 14.447825 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMSGRGKQ GGKARAKAKT RSSRAGLQFP VGRVHRLLRK GNYSERVGAG APVYLAAVLE YLTAEILELA GNAARDNKKT RIIPRHLQL AIRNDEELNK LLGRVTIAQG GVLPNIQAVL LPKKTESHHK AKGK UniProtKB: Histone H2A type 1-B/E |
-分子 #4: Histone H2B type 1-J
| 分子 | 名称: Histone H2B type 1-J / タイプ: protein_or_peptide / ID: 4 / コピー数: 2 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 14.217516 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GSHMPEPAKS APAPKKGSKK AVTKAQKKDG KKRKRSRKES YSIYVYKVLK QVHPDTGISS KAMGIMNSFV NDIFERIAGE ASRLAHYNK RSTITSREIQ TAVRLLLPGE LAKHAVSEGT KAVTKYTSAK UniProtKB: Histone H2B type 1-J |
-分子 #7: Histone acetyltransferase p300
| 分子 | 名称: Histone acetyltransferase p300 / タイプ: protein_or_peptide / ID: 7 / コピー数: 1 / 光学異性体: LEVO / EC番号: histone acetyltransferase |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 74.42657 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: TQSSPAPGQS KKKIFKPEEL RQALMPTLEA LYRQDPESLP FRQPVDPQLL GIPDYFDIVK SPMDLSTIKR KLDTGQYQEP WQYVDDIWL MFNNAWLYNR KTSRVYKYCS KLSEVFEQEI DPVMQSLGYC CGRKLEFSPQ TLCCYGKQLC TIPRDATYYS Y QNRYHFCE ...文字列: TQSSPAPGQS KKKIFKPEEL RQALMPTLEA LYRQDPESLP FRQPVDPQLL GIPDYFDIVK SPMDLSTIKR KLDTGQYQEP WQYVDDIWL MFNNAWLYNR KTSRVYKYCS KLSEVFEQEI DPVMQSLGYC CGRKLEFSPQ TLCCYGKQLC TIPRDATYYS Y QNRYHFCE KCFNEIQGES VSLGDDPSQP QTTINKEQFS KRKNDTLDPE LFVECTECGR KMHQICVLHH EIIWPAGFVC DG CLKKSAR TRKENKFSAK RLPSTRLGTF LENRVNDFLR RQNHPESGEV TVRVVHASDK TVEVKPGMKA RFVDSGEMAE SFP YRTKAL FAFEEIDGVD LCFFGMHVQE YGSDCPPPNQ RRVYISYLDS VHFFRPKCLR TAVYHEILIG YLEYVKKLGY TTGH IWACP PSEGDDYIFH CHPPDQKIPK PKRLQEWFKK MLDKAVSERI VHDYKDIFKQ ATEDRLTSAK ELPYFEGDFW PNVLE ESIK ESGGSGSQKL YATMEKHKEV FFVIRLIAGP AANSLPPIVD PDPLIPCDLM DGRDAFLTLA RDKHLEFSSL RRAQWS TMC MLVELHTQSQ DRFVYTCNEC KHHVETRWHC TVCEDYDLCI TCYNTKNHDH KMEKLGLGLD DESNNQQHHH HHHHH UniProtKB: Histone acetyltransferase p300, Histone acetyltransferase p300 |
-分子 #5: DNA (145-MER)
| 分子 | 名称: DNA (145-MER) / タイプ: dna / ID: 5 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種: synthetic construct (人工物) |
| 分子量 | 理論値: 44.520383 KDa |
| 配列 | 文字列: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT) ...文字列: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA) (DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC) (DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG) (DG)(DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT)(DC) (DT)(DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT)(DA) (DT)(DA)(DT)(DA)(DC)(DA) (DT)(DC)(DG) (DA)(DT) |
-分子 #6: DNA (145-MER)
| 分子 | 名称: DNA (145-MER) / タイプ: dna / ID: 6 / コピー数: 1 / 分類: DNA |
|---|---|
| 由来(天然) | 生物種: synthetic construct (人工物) |
| 分子量 | 理論値: 44.99166 KDa |
| 配列 | 文字列: (DA)(DT)(DC)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC) ...文字列: (DA)(DT)(DC)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC)(DC)(DC)(DC) (DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT)(DA) (DA)(DA) (DA)(DC)(DG)(DC)(DG)(DG)(DG) (DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC)(DG) (DT)(DA)(DC) (DG)(DT)(DG)(DC)(DG)(DT) (DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG) (DA)(DG)(DC)(DT)(DG) (DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC)(DA) (DA)(DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG) (DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC)(DC) (DG)(DG)(DG)(DA)(DT)(DT) (DC)(DT)(DG) (DA)(DT) |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 56.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD 最大 デフォーカス(公称値): 2.3000000000000003 µm 最小 デフォーカス(公称値): 1.2 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)