+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32365 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
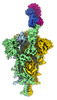
| Title | SARS-CoV-2 Delta S-8D3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 Delta variant spike protein / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / Attachment and Entry / entry receptor-mediated virion attachment to host cell / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / endocytosis involved in viral entry into host cell / receptor ligand activity / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell plasma membrane / SARS-CoV-2 activates/modulates innate and adaptive immune responses / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Cong Y / Liu CX | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Authors: Yifan Wang / Caixuan Liu / Chao Zhang / Yanxing Wang / Qin Hong / Shiqi Xu / Zuyang Li / Yong Yang / Zhong Huang / Yao Cong /  Abstract: The SARS-CoV-2 Delta variant is currently the dominant circulating strain in the world. Uncovering the structural basis of the enhanced transmission and altered immune sensitivity of Delta is ...The SARS-CoV-2 Delta variant is currently the dominant circulating strain in the world. Uncovering the structural basis of the enhanced transmission and altered immune sensitivity of Delta is particularly important. Here we present cryo-EM structures revealing two conformational states of Delta spike and S/ACE2 complex in four states. Our cryo-EM analysis suggests that RBD destabilizations lead to population shift towards the more RBD-up and S1 destabilized fusion-prone state, beneficial for engagement with ACE2 and shedding of S1. Noteworthy, we find the Delta T478K substitution plays a vital role in stabilizing and reshaping the RBM loop, enhancing interaction with ACE2. Collectively, increased propensity for more RBD-up states and the affinity-enhancing T478K substitution together contribute to increased ACE2 binding, providing structural basis of rapid spread of Delta. Moreover, we identify a previously generated MAb 8D3 as a cross-variant broadly neutralizing antibody and reveal that 8D3 binding induces a large K478 side-chain orientation change, suggesting 8D3 may use an "induced-fit" mechanism to tolerate Delta T478K mutation. We also find that all five RBD-targeting MAbs tested remain effective on Delta, suggesting that Delta well preserves the neutralizing antigenic landscape in RBD. Our findings shed new lights on the pathogenicity and antibody neutralization of Delta. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32365.map.gz emd_32365.map.gz | 17 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32365-v30.xml emd-32365-v30.xml emd-32365.xml emd-32365.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32365.png emd_32365.png | 53.1 KB | ||
| Filedesc metadata |  emd-32365.cif.gz emd-32365.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32365 http://ftp.pdbj.org/pub/emdb/structures/EMD-32365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32365 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32365 | HTTPS FTP |
-Related structure data
| Related structure data |  7w9eMC  7w92C  7w94C  7w98C  7w99C  7w9bC  7w9cC  7w9fC  7w9iC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32365.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32365.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.093 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : SARS-Cov2 Delta variant S-8D3
| Entire | Name: SARS-Cov2 Delta variant S-8D3 |
|---|---|
| Components |
|
-Supramolecule #1: SARS-Cov2 Delta variant S-8D3
| Supramolecule | Name: SARS-Cov2 Delta variant S-8D3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-Cov2 Delta variant S
| Supramolecule | Name: SARS-Cov2 Delta variant S / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: 8D3 Fab
| Supramolecule | Name: 8D3 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.085031 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLRT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFGV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNLRT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFGV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QT LLALHRS YLTPGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRV QPTESI VRFPNITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSF VIRGD EVRQIAPGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYRYRL FRKSNLKPFE RDISTEIYQA GSKPC NGVE GFNCYFPLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFL PFQ QFGRDIADTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ GVNCTEVPVA IHADQLTPTW RVYSTGS NV FQTRAGCLIG AEHVNNSYEC DIPIGAGICA SYQTQTNSRG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTI S VTTEILPVSM TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGF NFSQILPDPS KPSKRSFIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFG AGAALQIPFA MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STASALGKLQ NVVNQNAQAL N TLVKQLSS NFGAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RV DFCGKGY HLMSFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNT FVSGNC DVVIGIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDL QELGK YEQGSGYIPE APRDGQAYVR KDGEWVLLST FLENLYFQGD YKDDDDKHHH HHHHHH UniProtKB: Spike glycoprotein |
-Macromolecule #2: Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain
| Macromolecule | Name: Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.108055 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLQQSGPE LVKPGASVKI SCKTSGYTFT EYTMYWVKQS HGKSLEWIGG INPNIGDTSY NQNFKGKATL TVDRSSSTAY MELRSLTSE DSAVYYCARD GYPYYYALDH WGQGTSVTVS SAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS ...String: EVQLQQSGPE LVKPGASVKI SCKTSGYTFT EYTMYWVKQS HGKSLEWIGG INPNIGDTSY NQNFKGKATL TVDRSSSTAY MELRSLTSE DSAVYYCARD GYPYYYALDH WGQGTSVTVS SAKTTPPSVY PLAPGSAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS GVHTFPAVLQ SDLYTLSSSV TVPSSTWPSE TVTCNVAHPA SSTKVDKKIV PRDCGCKPCI CTVPEVSSVF IF PPKPKDV LTITLTPKVT CVVVDISKDD PEVQFSWFVD DVEVHTAQTQ PREEQFNSTF RSVSELPIMH QDWLNGKEFK CRV NSAAFP APIEKTISKT KGRPKAPQVY TIPPPKEQMA KDKVSLTCMI TDFFPEDITV EWQWNGQPAE NYKNTQPIMD TDGS YFVYS KLNVQKSNWE AGNTFTCSVL HEGLHNHHTE KSLSHSPGK |
-Macromolecule #3: The light chain of 8D3 fab
| Macromolecule | Name: The light chain of 8D3 fab / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.735051 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVMTQSQKF MSTSVGDRVS VTCKASQNVG TNVAWYQQKP GQSPKALIYS TSYRYSGVPD RFTGSGSGTD FTLTISNVQS EDLAEYFCQ QYNSYPYTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS ...String: DIVMTQSQKF MSTSVGDRVS VTCKASQNVG TNVAWYQQKP GQSPKALIYS TSYRYSGVPD RFTGSGSGTD FTLTISNVQS EDLAEYFCQ QYNSYPYTFG GGTKLEIKRA DAAPTVSIFP PSSEQLTSGG ASVVCFLNNF YPKDINVKWK IDGSERQNGV L NSWTDQDS KDSTYSMSST LTLTKDEYER HNSYTCEATH KTSTSPIVKS FNRNEC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 246240 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















