+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dimethylformamidase, 2x(A2B2) | |||||||||
 Map data Map data | B-factor sharpened map from post processing step in Relion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Amidohydrolase / mononuclear iron / AB polypeptide / tetramer / HYDROLASE | |||||||||
| Function / homology | N,N-dimethylformamidase / N,N-dimethylformamidase activity / N,N-dimethylformamidase beta subunit-like, C-terminal / N,N-dimethylformamidase beta subunit-like, C-terminal / Concanavalin A-like lectin/glucanase domain superfamily / metal ion binding / N,N-dimethylformamidase large subunit / N,N-dimethylformamidase small subunit Function and homology information Function and homology information | |||||||||
| Biological species |  Paracoccus sp. (bacteria) / Paracoccus sp. (bacteria) /  Paracoccus sp. SSG05 (bacteria) Paracoccus sp. SSG05 (bacteria) | |||||||||
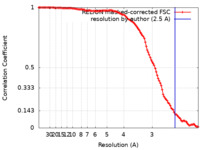
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Vinothkumar KR / Subramanian R | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Dimethylformamidase with a Unique Iron Center Authors: Arya C / Ramanathan G / Vinothkumar KR / Subramanian R | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32357.map.gz emd_32357.map.gz | 117.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32357-v30.xml emd-32357-v30.xml emd-32357.xml emd-32357.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32357_fsc.xml emd_32357_fsc.xml | 11.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_32357.png emd_32357.png | 71.8 KB | ||
| Filedesc metadata |  emd-32357.cif.gz emd-32357.cif.gz | 7.1 KB | ||
| Others |  emd_32357_half_map_1.map.gz emd_32357_half_map_1.map.gz emd_32357_half_map_2.map.gz emd_32357_half_map_2.map.gz | 97 MB 97.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32357 http://ftp.pdbj.org/pub/emdb/structures/EMD-32357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32357 | HTTPS FTP |
-Related structure data
| Related structure data |  7w8jMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32357.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32357.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map from post processing step in Relion | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: One of the half maps from final refinement
| File | emd_32357_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps from final refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: One of the half maps from final refinement
| File | emd_32357_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | One of the half maps from final refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DMFase, an amidohydrolase
| Entire | Name: DMFase, an amidohydrolase |
|---|---|
| Components |
|
-Supramolecule #1: DMFase, an amidohydrolase
| Supramolecule | Name: DMFase, an amidohydrolase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Paracoccus sp. (bacteria) / Strain: SSG05 Paracoccus sp. (bacteria) / Strain: SSG05 |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: N,N-dimethylformamidase large subunit
| Macromolecule | Name: N,N-dimethylformamidase large subunit / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: N,N-dimethylformamidase |
|---|---|
| Source (natural) | Organism:  Paracoccus sp. SSG05 (bacteria) Paracoccus sp. SSG05 (bacteria) |
| Molecular weight | Theoretical: 86.341758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKDIAIRGYC DRPSVATGET IRFYVSANET RGTFDAELVR LIHGDSNPAG PGYKEEAIKS DLEGQYPARF QRTQFGSYVE VADPDAGLQ PDGAFSVHLF LWSTTPSRGR QGIASRWNDE RQSGWNLAIE DGRVVFTIGD GSGATSSVVS DRPLFQQIWY S ITGVYDPE ...String: MKDIAIRGYC DRPSVATGET IRFYVSANET RGTFDAELVR LIHGDSNPAG PGYKEEAIKS DLEGQYPARF QRTQFGSYVE VADPDAGLQ PDGAFSVHLF LWSTTPSRGR QGIASRWNDE RQSGWNLAIE DGRVVFTIGD GSGATSSVVS DRPLFQQIWY S ITGVYDPE KKQLRLYQKS VVNRTNSRFG LVVPLDSDCA VSADATVKAA DSETSLLIAG LGEAAAQDGR TWCIAHYNGK VD APKIYGC ALGQDDAEKL SRGEIVRPIS RLAHWDFSAG IGLNGIPTDH VVDASGYGHH GRCMNQPSRG STGWNWDGHE ENF IHCPEQ YGALWFHEDC LDDCRWEKDF EFTVPEGLKS DFYAVKIRYE DTEDYIPFFV LPPRGTATAP ILVIASTLSY LAYA NEQIM HKADIGQAVA GHTPVLNEND VELHKNLSYY GLSTYDGHID GRGVQYTSWR RPIMNLRPKH RQGFGSIWEL PADLH LIDW LNHNGFEYDV ATEHDLNDQG AELLRRYKVV LTGSHPEYQT WANADAWEDY LADGGRGMYL AANGMYWIVE VHPEKP WVM EVRKELGVTA WEAPPGEYHY STNGRRGGRF RGRARATQKI WGTGMSSFGF DHSGYFVQMP DSQDERVAWI MEGIDPE ER IGDGGLVGGG AGGYELDRYD LALGTPPNTL LLASSVEHSV VYTVIPDDKA FPHPGMNGGE HPFVRADITY FSTANGGG M FATSSISWLG SLSWNDYDNN VSKMTKNVLN QFIKDEPAPR VKLAAALEHH HHHH UniProtKB: N,N-dimethylformamidase large subunit |
-Macromolecule #2: N,N-dimethylformamidase small subunit
| Macromolecule | Name: N,N-dimethylformamidase small subunit / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO / EC number: N,N-dimethylformamidase |
|---|---|
| Source (natural) | Organism:  Paracoccus sp. SSG05 (bacteria) Paracoccus sp. SSG05 (bacteria) |
| Molecular weight | Theoretical: 16.083823 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTEASESCVR DPSNYRDRSA DWYAFYDERR RKEIIDIIDE HPEIVEEHAA NPFGYRKHPS PYLQRVHNYF RMQPTFGRYY IYSEREWDA YRIATIREFG ELPELGDERF KTEEEAMHAV FLRRIEDVRA ELA UniProtKB: N,N-dimethylformamidase small subunit |
-Macromolecule #3: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #4: water
| Macromolecule | Name: water / type: ligand / ID: 4 / Number of copies: 290 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: wait time - 10 seconds and 3.5 seconds blot. | |||||||||
| Details | Enzyme after gel filtration was used for cryoEM grid prep. Sample was held at 37 degrees before application on the grid. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.0 K |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 548 / Average exposure time: 60.0 sec. / Average electron dose: 28.3 e/Å2 Details: One image was collected per hole. Total of 25 frames was saved and each frame has ~1.13 e/A2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 130841 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 0.85 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Servalcat was used along with Refmac to calculate fofc maps and extensively used for modelling. | ||||||
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 73.98 | ||||||
| Output model |  PDB-7w8j: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)