+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human NaV1.3/beta1/beta2-ICA121431 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / Drug / antagonist / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during Purkinje myocyte cell action potential / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / cardiac conduction / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential ...response to pyrethroid / corticospinal neuron axon guidance / positive regulation of voltage-gated sodium channel activity / voltage-gated sodium channel activity involved in cardiac muscle cell action potential / regulation of atrial cardiac muscle cell membrane depolarization / membrane depolarization during Purkinje myocyte cell action potential / voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization / cardiac conduction / membrane depolarization during cardiac muscle cell action potential / membrane depolarization during action potential / positive regulation of sodium ion transport / regulation of sodium ion transmembrane transport / axon initial segment / regulation of ventricular cardiac muscle cell membrane repolarization / cardiac muscle cell action potential involved in contraction / node of Ranvier / voltage-gated sodium channel complex / sodium channel inhibitor activity / locomotion / neuronal action potential propagation / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / sodium ion transport / Phase 0 - rapid depolarisation / regulation of heart rate by cardiac conduction / behavioral response to pain / intercalated disc / sarcoplasm / membrane depolarization / sodium channel regulator activity / cardiac muscle contraction / T-tubule / axon guidance / basal plasma membrane / sodium ion transmembrane transport / positive regulation of neuron projection development / Sensory perception of sweet, bitter, and umami (glutamate) taste / nervous system development / response to heat / perikaryon / gene expression / chemical synaptic transmission / transmembrane transporter binding / cell adhesion / synapse / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||
 Authors Authors | Jiang D / Li X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for modulation of human Na1.3 by clinical drug and selective antagonist. Authors: Xiaojing Li / Feng Xu / Hao Xu / Shuli Zhang / Yiwei Gao / Hongwei Zhang / Yanli Dong / Yanchun Zheng / Bei Yang / Jianyuan Sun / Xuejun Cai Zhang / Yan Zhao / Daohua Jiang /   Abstract: Voltage-gated sodium (Na) channels play fundamental roles in initiating and propagating action potentials. Na1.3 is involved in numerous physiological processes including neuronal development, ...Voltage-gated sodium (Na) channels play fundamental roles in initiating and propagating action potentials. Na1.3 is involved in numerous physiological processes including neuronal development, hormone secretion and pain perception. Here we report structures of human Na1.3/β1/β2 in complex with clinically-used drug bulleyaconitine A and selective antagonist ICA121431. Bulleyaconitine A is located around domain I-II fenestration, providing the detailed view of the site-2 neurotoxin binding site. It partially blocks ion path and expands the pore-lining helices, elucidating how the bulleyaconitine A reduces peak amplitude but improves channel open probability. In contrast, ICA121431 preferentially binds to activated domain IV voltage-sensor, consequently strengthens the Ile-Phe-Met motif binding to its receptor site, stabilizes the channel in inactivated state, revealing an allosterically inhibitory mechanism of Na channels. Our results provide structural details of distinct small-molecular modulators binding sites, elucidate molecular mechanisms of their action on Na channels and pave a way for subtype-selective therapeutic development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32343.map.gz emd_32343.map.gz | 59.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32343-v30.xml emd-32343-v30.xml emd-32343.xml emd-32343.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32343.png emd_32343.png | 111.3 KB | ||
| Filedesc metadata |  emd-32343.cif.gz emd-32343.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32343 http://ftp.pdbj.org/pub/emdb/structures/EMD-32343 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32343 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32343 | HTTPS FTP |
-Validation report
| Summary document |  emd_32343_validation.pdf.gz emd_32343_validation.pdf.gz | 723.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32343_full_validation.pdf.gz emd_32343_full_validation.pdf.gz | 722.9 KB | Display | |
| Data in XML |  emd_32343_validation.xml.gz emd_32343_validation.xml.gz | 6.5 KB | Display | |
| Data in CIF |  emd_32343_validation.cif.gz emd_32343_validation.cif.gz | 7.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32343 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32343 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32343 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32343 | HTTPS FTP |
-Related structure data
| Related structure data |  7w7fMC  7w77C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32343.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32343.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
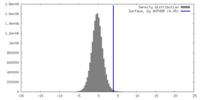
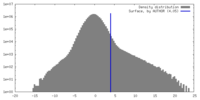
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Sodium channel complex of type 3 subunit alpha with auxiliary bet...
| Entire | Name: Sodium channel complex of type 3 subunit alpha with auxiliary beta-1 and beta-2 |
|---|---|
| Components |
|
-Supramolecule #1: Sodium channel complex of type 3 subunit alpha with auxiliary bet...
| Supramolecule | Name: Sodium channel complex of type 3 subunit alpha with auxiliary beta-1 and beta-2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Sodium channel subunit beta-1
| Macromolecule | Name: Sodium channel subunit beta-1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.732115 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV ...String: MGRLLALVVG AALVSSACGG CVEVDSETEA VYGMTFKILC ISCKRRSETN AETFTEWTFR QKGTEEFVKI LRYENEVLQL EEDERFEGR VVWNGSRGTK DLQDLSIFIT NVTYNHSGDY ECHVYRLLFF ENYEHNTSVV KKIHIEVVDK ANRDMASIVS E IMMYVLIV VLTIWLVAEM IYCYKKIAAA TETAAQENAS EYLAITSESK ENCTGVQVAE UniProtKB: Sodium channel regulatory subunit beta-1 |
-Macromolecule #2: Sodium channel subunit beta-2
| Macromolecule | Name: Sodium channel subunit beta-2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.355859 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHRDAWLPRP AFSLTGLSLF FSLVPPGRSM EVTVPATLNV LNGSDARLPC TFNSCYTVNH KQFSLNWTYQ ECNNCSEEMF LQFRMKIIN LKLERFQDRV EFSGNPSKYD VSVMLRNVQP EDEGIYNCYI MNPPDRHRGH GKIHLQVLME EPPERDSTVA V IVGASVGG ...String: MHRDAWLPRP AFSLTGLSLF FSLVPPGRSM EVTVPATLNV LNGSDARLPC TFNSCYTVNH KQFSLNWTYQ ECNNCSEEMF LQFRMKIIN LKLERFQDRV EFSGNPSKYD VSVMLRNVQP EDEGIYNCYI MNPPDRHRGH GKIHLQVLME EPPERDSTVA V IVGASVGG FLAVVILVLM VVKCVRRKKE QKLSTDDLKT EEEGKTDGEG NPDDGAK UniProtKB: Sodium channel regulatory subunit beta-2 |
-Macromolecule #3: Sodium channel protein type 3 subunit alpha
| Macromolecule | Name: Sodium channel protein type 3 subunit alpha / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 226.516453 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAQALLVPPG PESFRLFTRE SLAAIEKRAA EEKAKKPKKE QDNDDENKPK PNSDLEAGKN LPFIYGDIPP EMVSEPLEDL DPYYINKKT FIVMNKGKAI FRFSATSALY ILTPLNPVRK IAIKILVHSL FSMLIMCTIL TNCVFMTLSN PPDWTKNVEY T FTGIYTFE ...String: MAQALLVPPG PESFRLFTRE SLAAIEKRAA EEKAKKPKKE QDNDDENKPK PNSDLEAGKN LPFIYGDIPP EMVSEPLEDL DPYYINKKT FIVMNKGKAI FRFSATSALY ILTPLNPVRK IAIKILVHSL FSMLIMCTIL TNCVFMTLSN PPDWTKNVEY T FTGIYTFE SLIKILARGF CLEDFTFLRD PWNWLDFSVI VMAYVTEFVS LGNVSALRTF RVLRALKTIS VIPGLKTIVG AL IQSVKKL SDVMILTVFC LSVFALIGLQ LFMGNLRNKC LQWPPSDSAF ETNTTSYFNG TMDSNGTFVN VTMSTFNWKD YIG DDSHFY VLDGQKDPLL CGNGSDAGQC PEGYICVKAG RNPNYGYTSF DTFSWAFLSL FRLMTQDYWE NLYQLTLRAA GKTY MIFFV LVIFLGSFYL VNLILAVVAM AYEEQNQATL EEAEQKEAEF QQMLEQLKKQ QEEAQAVAAA SAASRDFSGI GGLGE LLES SSEASKLSSK SAKEWRNRRK KRRQREHLEG NNKGERDSFP KSESEDSVKR SSFLFSMDGN RLTSDKKFCS PHQSLL SIR GSLFSPRRNS KTSIFSFRGR AKDVGSENDF ADDEHSTFED SESRRDSLFV PHRHGERRNS NVSQASMSSR MVPGLPA NG KMHSTVDCNG VVSLVGGPSA LTSPTGQLPP EGTTTETEVR KRRLSSYQIS MEMLEDSSGR QRAVSIASIL TNTMEELE E SRQKCPPCWY RFANVFLIWD CCDAWLKVKH LVNLIVMDPF VDLAITICIV LNTLFMAMEH YPMTEQFSSV LTVGNLVFT GIFTAEMVLK IIAMDPYYYF QEGWNIFDGI IVSLSLMELG LSNVEGLSVL RSFRLLRVFK LAKSWPTLNM LIKIIGNSVG ALGNLTLVL AIIVFIFAVV GMQLFGKSYK ECVCKINDDC TLPRWHMNDF FHSFLIVFRV LCGEWIETMW DCMEVAGQTM C LIVFMLVM VIGNLVVLNL FLALLLSSFS SDNLAATDDD NEMNNLQIAV GRMQKGIDYV KNKMRECFQK AFFRKPKVIE IH EGNKIDS CMSNNTGIEI SKELNYLRDG NGTTSGVGTG SSVEKYVIDE NDYMSFINNP SLTVTVPIAV GESDFENLNT EEF SSESEL EESKEKLNAT SSSEGSTVDV VLPREGEQAE TEPEEDLKPE ACFTEGCIKK FPFCQVSTEE GKGKIWWNLR KTCY SIVEH NWFETFIVFM ILLSSGALAF EDIYIEQRKT IKTMLEYADK VFTYIFILEM LLKWVAYGFQ TYFTNAWCWL DFLIV DVSL VSLVANALGY SELGAIKSLR TLRALRPLRA LSRFEGMRVV VNALVGAIPS IMNVLLVCLI FWLIFSIMGV NLFAGK FYH CVNMTTGNMF DISDVNNLSD CQALGKQARW KNVKVNFDNV GAGYLALLQV ATFKGWMDIM YAAVDSRDVK LQPVYEE NL YMYLYFVIFI IFGSFFTLNL FIGVIIDNFN QQKKKFGGQD IFMTEEQKKY YNAMKKLGSK KPQKPIPRPA NKFQGMVF D FVTRQVFDIS IMILICLNMV TMMVETDDQG KYMTLVLSRI NLVFIVLFTG EFVLKLVSLR HYYFTIGWNI FDFVVVILS IVGMFLAEMI EKYFVSPTLF RVIRLARIGR ILRLIKGAKG IRTLLFALMM SLPALFNIGL LLFLVMFIYA IFGMSNFAYV KKEAGIDDM FNFETFGNSM ICLFQITTSA GWDGLLAPIL NSAPPDCDPD TIHPGSSVKG DCGNPSVGIF FFVSYIIISF L VVVNMYIA VILENFSVAT EESAEPLSED DFEMFYEVWE KFDPDATQFI EFSKLSDFAA ALDPPLLIAK PNKVQLIAMD LP MVSGDRI HCLDILFAFT KRVLGESGEM DALRIQMEDR FMASNPSKVS YEPITTTLKR KQEEVSAAII QRNFRCYLLK QRL KNISSN YNKEAIKGRI DLPIKQDMII DKLNGNSTPE KTDGSSSTTS PPSYDSVTKP DKEKFEKDKP EKESKGKEVR ENQK UniProtKB: Sodium channel protein type 3 subunit alpha |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyl...
| Macromolecule | Name: [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate type: ligand / ID: 7 / Number of copies: 17 / Formula: 6OU |
|---|---|
| Molecular weight | Theoretical: 717.996 Da |
| Chemical component information |  ChemComp-6OU: |
-Macromolecule #8: 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide
| Macromolecule | Name: 2,2-diphenyl-~{N}-[4-(1,3-thiazol-2-ylsulfamoyl)phenyl]ethanamide type: ligand / ID: 8 / Number of copies: 1 / Formula: 8DE |
|---|---|
| Molecular weight | Theoretical: 449.545 Da |
| Chemical component information |  ChemComp-8DE: |
-Macromolecule #9: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]sp...
| Macromolecule | Name: (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en type: ligand / ID: 9 / Number of copies: 1 / Formula: 9Z9 |
|---|---|
| Molecular weight | Theoretical: 544.805 Da |
| Chemical component information |  ChemComp-9Z9: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)