+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3232 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The architecture of the S. pombe CCR4-NOT complex | |||||||||
 Map data Map data | Cryo-EM reconstruction of the CCR4-NOT complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CCR4-NOT / cryo-electron microscopy / RNA decay / deadenylation / single-particle 3D reconstruction | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Ukleja M / Cuellar J / Siwaszek A / Kasprzak JM / Czarnocki-Cieciura M / Bujnicki J / Dziembowski A / Valpuesta JM | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: The architecture of the Schizosaccharomyces pombe CCR4-NOT complex. Authors: Marta Ukleja / Jorge Cuellar / Aleksandra Siwaszek / Joanna M Kasprzak / Mariusz Czarnocki-Cieciura / Janusz M Bujnicki / Andrzej Dziembowski / Jose M Valpuesta /   Abstract: CCR4-NOT is a large protein complex present both in cytoplasm and the nucleus of eukaryotic cells. Although it is involved in a variety of distinct processes related to expression of genetic ...CCR4-NOT is a large protein complex present both in cytoplasm and the nucleus of eukaryotic cells. Although it is involved in a variety of distinct processes related to expression of genetic information such as poly(A) tail shortening, transcription regulation, nuclear export and protein degradation, there is only fragmentary information available on some of its nine subunits. Here we show a comprehensive structural characterization of the native CCR4-NOT complex from Schizosaccharomyces pombe. Our cryo-EM 3D reconstruction of the complex, combined with techniques such as immunomicroscopy, RNA-nanogold labelling, docking of the available high-resolution structures and models of different subunits and domains, allow us to propose its full molecular architecture. We locate all functionally defined domains endowed with deadenylating and ubiquitinating activities, the nucleus-specific RNA-interacting subunit Mmi1, as well as surfaces responsible for protein-protein interactions. This information provides insight into cooperation of the different CCR4-NOT complex functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3232.map.gz emd_3232.map.gz | 1.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3232-v30.xml emd-3232-v30.xml emd-3232.xml emd-3232.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3232_ccr4notCryoEM500x500.jpg EMD-3232_ccr4notCryoEM500x500.jpg | 65.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3232 http://ftp.pdbj.org/pub/emdb/structures/EMD-3232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3232 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3232 | HTTPS FTP |
-Validation report
| Summary document |  emd_3232_validation.pdf.gz emd_3232_validation.pdf.gz | 182.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3232_full_validation.pdf.gz emd_3232_full_validation.pdf.gz | 181.5 KB | Display | |
| Data in XML |  emd_3232_validation.xml.gz emd_3232_validation.xml.gz | 4.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3232 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3232 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3232 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3232.map.gz / Format: CCP4 / Size: 1.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3232.map.gz / Format: CCP4 / Size: 1.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM reconstruction of the CCR4-NOT complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
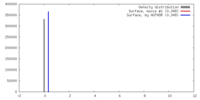
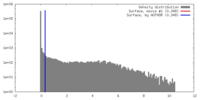
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : S. pombe CCR4-NOT complex
| Entire | Name: S. pombe CCR4-NOT complex |
|---|---|
| Components |
|
-Supramolecule #1000: S. pombe CCR4-NOT complex
| Supramolecule | Name: S. pombe CCR4-NOT complex / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 700 MDa |
-Macromolecule #1: CCR4-NOT complex
| Macromolecule | Name: CCR4-NOT complex / type: protein_or_peptide / ID: 1 Details: CCR4-NOT complex purified from the endogenous expression of S. pombe cells. The complex is composed of Not1, Not2, Not3, Not4, Caf1, Caf40, Ccr4, Mmi1 subunits Number of copies: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 700 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 10 mM HEPES, 500 mM NaCl, ~3% glycerol |
| Grid | Details: Quantifoil R 1.2/ R1.3 300 mesh grids; ref. Q09684 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: LEICA EM CPC Method: Aliquots (5 ul) of purified concentrated CCR4-NOT were incubated (2-5 min) with the grid, blotted and plunged into a liquid ethane chamber. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Date | May 20, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI EAGLE (4k x 4k) / Digitization - Sampling interval: 4.2 µm / Number real images: 600 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.0 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | single-particle 3D reconstruction was performed using XMIPP 2.4 and EMAN1, CTFFIND3 for the CTF determination |
|---|---|
| CTF correction | Details: CTFIND3, all micrograph |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Software - Name: XMIPP, 2.4, EMAN1 / Number images used: 20500 |
| Final two d classification | Number classes: 200 |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)