[English] 日本語
 Yorodumi
Yorodumi- EMDB-32298: Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of Human Gastrin Releasing Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | GPCR / Gastrin Releasing Peptide Receptor / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to external biotic stimulus / positive regulation of respiratory gaseous exchange / positive regulation of behavioral fear response / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / neuropeptide receptor activity / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of platelet activation ...response to external biotic stimulus / positive regulation of respiratory gaseous exchange / positive regulation of behavioral fear response / Fatty Acids bound to GPR40 (FFAR1) regulate insulin secretion / Acetylcholine regulates insulin secretion / phospholipase C-activating G protein-coupled glutamate receptor signaling pathway / neuropeptide receptor activity / PLC beta mediated events / phospholipase C-activating serotonin receptor signaling pathway / regulation of platelet activation / entrainment of circadian clock / psychomotor behavior / neuropeptide binding / G protein-coupled peptide receptor activity / regulation of canonical Wnt signaling pathway / glutamate receptor signaling pathway / motor behavior / phototransduction, visible light / carbohydrate transmembrane transporter activity / social behavior / photoreceptor outer segment / neuropeptide signaling pathway / postsynaptic cytosol / enzyme regulator activity / response to prostaglandin E / GTPase activator activity / Peptide ligand-binding receptors / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / blood coagulation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / Inactivation, recovery and regulation of the phototransduction cascade / G alpha (12/13) signalling events / regulation of cell population proliferation / extracellular vesicle / sensory perception of taste / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / G protein activity / retina development in camera-type eye / GTPase binding / Ca2+ pathway / fibroblast proliferation / nuclear membrane / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Ras protein signal transduction / learning or memory / periplasmic space / Extra-nuclear estrogen signaling / cell population proliferation / protein stabilization / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / GTP binding / protein-containing complex binding / Golgi apparatus / signal transduction / extracellular exosome / metal ion binding / membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||||||||||||||
 Authors Authors | Zhan Y / Peng S | |||||||||||||||||||||
| Funding support |  China, 6 items China, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structures of human gastrin-releasing peptide receptors bound to antagonist and agonist for cancer and itch therapy. Authors: Shuman Peng / Yuting Zhan / Dongqi Zhang / Lu Ren / Anqi Chen / Zhou-Feng Chen / Haitao Zhang /   Abstract: Gastrin releasing peptide receptor (GRPR), a member of the bombesin (BBN) G protein-coupled receptors, is aberrantly overexpressed in several malignant tumors, including those of the breast, ...Gastrin releasing peptide receptor (GRPR), a member of the bombesin (BBN) G protein-coupled receptors, is aberrantly overexpressed in several malignant tumors, including those of the breast, prostate, pancreas, lung, and central nervous system. Additionally, it also mediates non-histaminergic itch and pathological itch conditions in mice. Thus, GRPR could be an attractive target for cancer and itch therapy. Here, we report the inactive state crystal structure of human GRPR in complex with the non-peptide antagonist PD176252, as well as two active state cryo-electron microscopy (cryo-EM) structures of GRPR bound to the endogenous peptide agonist gastrin-releasing peptide and the synthetic BBN analog [D-Phe, β-Ala, Phe, Nle] Bn (6-14), in complex with G heterotrimers. These structures revealed the molecular mechanisms for the ligand binding, receptor activation, and G proteins signaling of GRPR, which are expected to accelerate the structure-based design of GRPR antagonists and agonists for the treatments of cancer and pruritus. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32298.map.gz emd_32298.map.gz | 4.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32298-v30.xml emd-32298-v30.xml emd-32298.xml emd-32298.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32298.png emd_32298.png | 24.3 KB | ||
| Filedesc metadata |  emd-32298.cif.gz emd-32298.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32298 http://ftp.pdbj.org/pub/emdb/structures/EMD-32298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32298 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32298 | HTTPS FTP |
-Related structure data
| Related structure data |  7w40MC  7w3zC  7w41C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32298.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32298.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
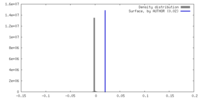
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Peptide Receptor in complex with the agonist Bombesin (6-14) [D-P...
| Entire | Name: Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers and ScFv16 |
|---|---|
| Components |
|
-Supramolecule #1: Peptide Receptor in complex with the agonist Bombesin (6-14) [D-P...
| Supramolecule | Name: Peptide Receptor in complex with the agonist Bombesin (6-14) [D-Phe6, beta-Ala11, Phe13, Nle14] and Gq heterotrimers and ScFv16 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Maltodextrin-binding protein,Gastrin-releasing peptide receptor
| Macromolecule | Name: Maltodextrin-binding protein,Gastrin-releasing peptide receptor type: protein_or_peptide / ID: 1 Details: The fusion protein of Maltodextrin-binding, protein,HiBit expression tag and Gastrin-releasing peptide receptor Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.534102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHENLYFQ GKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFP QVAATGDGPD IIFWAHDRFG GYAQSGLLAE ITPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN K DLLPNPPK ...String: MKTIIALSYI FCLVFADYKD DDDKHHHHHH HHHHENLYFQ GKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFP QVAATGDGPD IIFWAHDRFG GYAQSGLLAE ITPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN K DLLPNPPK TWEEIPALDK ELKAKGKSAL MFNLQEPYFT WPLIAADGGY AFKYAAGKYD IKDVGVDNAG AKAGLTFLVD LI KNKHMNA DTDYSIAEAA FNKGETAMTI NGPWAWSNID TSAVNYGVTV LPTFKGQPSK PFVGVLSAGI NAASPNKELA KEF LENYLL TDEGLEAVNK DKPLGAVALK SYEEELAKDP RIAATMENAQ KGEIMPNIPQ MSAFWYAVRT AVINAASGRQ TVDE ALKDA QTHSADLPVN DDWSHPGILY VIPAVYGVII LIGLIGNITL IKIFCTVKSM RNVPNLFISS LALGDLLLLI TCAPV DASR YLADRWLFGR IGCKLIPFIQ LTSVGVSVFT LTALSADRYK AIVRPMDIQA SHALMKACLK AAFIWIISML LAIPEA VFS DLHPFHEEST NQTFISCAPY PHSNELHPKI HSMASFLVFY VIPLSIISVY YYFIAKNLIQ SAYNLPVEGN IHVKKQI ES RKRLAKTVLV FVGLFAFCWL PNHVIYLYRS YHYSEVDTSM LHFVTSICAR LLAFTNSCVN PFALYLLSKS FRKQFNTQ L LCCQGSSGGG GSGGGGSSGF TLEDFVGDWE QTAAYNLDQV LEQGGVSSLL QNLAVSVTPI QRIVRSGENA LKIDIHVII PYEGLSADQM AQIEEVFKVV YPVDDHHFKV ILPYGTLVID GVTPNMLNYF GRPYEGIAVF DGKKITVTGT LWNGNKIIDE RLITPDGSM LFRVTINS UniProtKB: Maltodextrin-binding protein, Gastrin-releasing peptide receptor |
-Macromolecule #2: Guanine nucleotide-binding protein G(q) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.194844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA RRELKLLLLG TGESGKSTFI KQMRIIHGSG YSDEDKRGFT KLVYQNIFTA MQAMIRAMD TLKIPYKYEH NKAHAQLVRE VDVEKVSAFE NPYVDAIKSL WNDPGIQECY DRRREYQLSD STKYYLNDLD R VADPAYLP ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA RRELKLLLLG TGESGKSTFI KQMRIIHGSG YSDEDKRGFT KLVYQNIFTA MQAMIRAMD TLKIPYKYEH NKAHAQLVRE VDVEKVSAFE NPYVDAIKSL WNDPGIQECY DRRREYQLSD STKYYLNDLD R VADPAYLP TQQDVLRVQV PTTGIIEYPF DLQSVIFRMV DVGGLRSERR KWIHCFENVT SIMFLVALSE YDQVLVESDN EN RMEESKA LFRTIITYPW FQNSSVILFL NKKDLLEEKI MYSHLVDYFP EYDGPQRDAQ AAREFILKMF VDLNPDSDKI IYS HFTCAT DTENIRFVFA AVKDTILQLN LKEYNLV UniProtKB: Guanine nucleotide-binding protein G(q) subunit alpha |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.398137 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HGSSGSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQ DGKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY L SCCRFLDD ...String: MHHHHHHHHH HGSSGSELDQ LRQEAEQLKN QIRDARKACA DATLSQITNN IDPVGRIQMR TRRTLRGHLA KIYAMHWGTD SRLLVSASQ DGKLIIWDSY TTNKVHAIPL RSSWVMTCAY APSGNYVACG GLDNICSIYN LKTREGNVRV SRELAGHTGY L SCCRFLDD NQIVTSSGDT TCALWDIETG QQTTTFTGHT GDVMSLSLAP DTRLFVSGAC DASAKLWDVR EGMCRQTFTG HE SDINAIC FFPNGNAFAT GSDDATCRLF DLRADQELMT YSHDNIICGI TSVSFSKSGR LLLAGYDDFN CNVWDALKAD RAG VLAGHD NRVSCLGVTD DGMAVATGSW DSFLKIWNGS SGGGGSGGGG SSGVSGWRLF KKIS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.242612 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: ScFv16
| Macromolecule | Name: ScFv16 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.785582 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFADV QLVESGGGLV QPGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSEDT AMYYCVRSIY YYGSSPFDFW GQGTTLTVSS G GGGSGGGG ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFADV QLVESGGGLV QPGGSRKLSC SASGFAFSSF GMHWVRQAPE KGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSEDT AMYYCVRSIY YYGSSPFDFW GQGTTLTVSS G GGGSGGGG SGGGGSDIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLELKE NLYFQGHHHH HHHHHH |
-Macromolecule #6: Bombesin
| Macromolecule | Name: Bombesin / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 1.118307 KDa |
| Sequence | String: (DPN)QWAV(BAL)HF(NLN) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 BASE (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)