+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of McrBC (stalkless mutant) | |||||||||
 Map data Map data | This is the B-factor sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ protein / GTPase / endonuclease / McrBC / stalkless mutant / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtype IV site-specific deoxyribonuclease activity / restriction endodeoxyribonuclease activity / endonuclease complex / double-stranded methylated DNA binding / hemi-methylated DNA-binding / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / DNA restriction-modification system / DNA catabolic process / endonuclease activity / GTPase activity ...type IV site-specific deoxyribonuclease activity / restriction endodeoxyribonuclease activity / endonuclease complex / double-stranded methylated DNA binding / hemi-methylated DNA-binding / Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters / DNA restriction-modification system / DNA catabolic process / endonuclease activity / GTPase activity / GTP binding / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
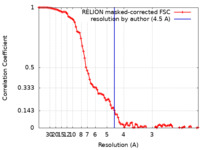
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Saikrishnan K / Adhav VA | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of McrBC (stalkless mutant) Authors: Saikrishnan K / Adhav VA / Bose S / Kutti R V #1:  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Structure-based mechanism for activation of the AAA+ GTPase McrB by the endonuclease McrC. Authors: Nirwan N / Itoh Y / Singh P / Bandyopadhyay S / Vinothkumar KR / Amunts A / Saikrishnan K | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32114.map.gz emd_32114.map.gz | 58.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32114-v30.xml emd-32114-v30.xml emd-32114.xml emd-32114.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32114_fsc.xml emd_32114_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_32114.png emd_32114.png | 84.4 KB | ||
| Filedesc metadata |  emd-32114.cif.gz emd-32114.cif.gz | 7 KB | ||
| Others |  emd_32114_half_map_1.map.gz emd_32114_half_map_1.map.gz emd_32114_half_map_2.map.gz emd_32114_half_map_2.map.gz | 48.4 MB 48.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32114 http://ftp.pdbj.org/pub/emdb/structures/EMD-32114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32114 | HTTPS FTP |
-Related structure data
| Related structure data |  7vsrMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32114.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32114.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the B-factor sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: This is the unfiltered half-map 2
| File | emd_32114_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the unfiltered half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
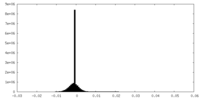
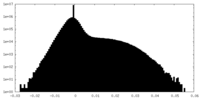
| Density Histograms |
-Half map: This is the unfiltered half-map 1
| File | emd_32114_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the unfiltered half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
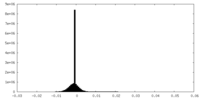
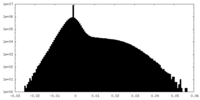
| Density Histograms |
- Sample components
Sample components
-Entire : McrBC (stalkless mutant)
| Entire | Name: McrBC (stalkless mutant) |
|---|---|
| Components |
|
-Supramolecule #1: McrBC (stalkless mutant)
| Supramolecule | Name: McrBC (stalkless mutant) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 720 KDa |
-Macromolecule #1: 5-methylcytosine-specific restriction enzyme B
| Macromolecule | Name: 5-methylcytosine-specific restriction enzyme B / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO EC number: Hydrolases; Acting on ester bonds; Endodeoxyribonucleases producing 5'-phosphomonoesters |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.272113 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MESIQPWIEK FIKQAQQQRS QSTKDYPTSY RNLRVKLSFG YGNFTSIPWF AFLGEGQEAS NGIYPVILYY KDFDELVLAY GISDTNEPH AQWQFSSDIP KTIAEYFQAT SGVYPKKYGQ SYYACSQKVS QGIDYTRFAS MLDNIINDYK LIFNSGKSVI P PMSKTESY ...String: MESIQPWIEK FIKQAQQQRS QSTKDYPTSY RNLRVKLSFG YGNFTSIPWF AFLGEGQEAS NGIYPVILYY KDFDELVLAY GISDTNEPH AQWQFSSDIP KTIAEYFQAT SGVYPKKYGQ SYYACSQKVS QGIDYTRFAS MLDNIINDYK LIFNSGKSVI P PMSKTESY CLEDALNDLF IPETTIETIL KRLTIKKNII LQGPPGVGKT FVARRLAYLL TGEKAPQRVN MVQFHQSYSY ED FIQGYRP NGVGFRRKDG IFYNFCQQAK EQPEKKYIFI IDEINRANLS KVFGEVMMLM EHDKRGENWS VPLTYSENDE ERF YVPENV YIIGLMNTAD RSLAVVDYAL RRRFSFIDIE PGFDTPQFRN FLLNKKAEPS FVESLCQKMN ELNQEISKEA TILG KGFRI GHSYFCCGLE DGTSPDTQWL NEIVMTDIAP LLEEYFFDDP YKQQKWTNKL LGDSSGSHHH HHH UniProtKB: Type IV methyl-directed restriction enzyme EcoKMcrB subunit |
-Macromolecule #2: Protein McrC
| Macromolecule | Name: Protein McrC / type: protein_or_peptide / ID: 2 Details: Residues from 60 to 99 in database P15006 are replaced by GG Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.22352 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEQPVIPVRN IYYMLTYAWG YLQEIKQANL EAIPGNNLLD ILGYVLNKGV LQLSRRGLEG GNEDTLANRI IKSTLAILIK HEKLNSTIR DEARSLYRKL PGISTLHLTP QHFSYLNGGK NTRYYKFVIS VCKFIVNNSI PGQNKGHYRF YDFERNEKEM S LLYQKFLY ...String: MEQPVIPVRN IYYMLTYAWG YLQEIKQANL EAIPGNNLLD ILGYVLNKGV LQLSRRGLEG GNEDTLANRI IKSTLAILIK HEKLNSTIR DEARSLYRKL PGISTLHLTP QHFSYLNGGK NTRYYKFVIS VCKFIVNNSI PGQNKGHYRF YDFERNEKEM S LLYQKFLY EFCRRELTSA NTTRSYLKWD ASSISDQSLN LLPRMETDIT IRSSEKILIV DAKYYKSIFS RRMGTEKFHS QN LYQLMNY LWSLKPENGE NIGGLLIYPH VDTAVKHRYK INGFDIGLCT VNLGQEWPCI HQELLDIFDE YLK UniProtKB: Type IV methyl-directed restriction enzyme EcoKMcrBC, Type IV methyl-directed restriction enzyme EcoKMcrBC |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER / type: ligand / ID: 3 / Number of copies: 6 / Formula: GNP |
|---|---|
| Molecular weight | Theoretical: 522.196 Da |
| Chemical component information |  ChemComp-GNP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R3.5/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR Details: The grid was floated with carbon before glow discharge | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 289 K / Instrument: FEI VITROBOT MARK IV Details: The sample was blot for 3.0 sec before plunging into liquid ethane. | |||||||||||||||
| Details | The McrBC complex was incubated with 2.5mM of GMP-PNP in the presence of 10mM Tris-Cl pH 8.0, 50mM KCl, 5mM MgCl2, 1mM DTT |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 2071 / Average exposure time: 60.0 sec. / Average electron dose: 27.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)