[English] 日本語
 Yorodumi
Yorodumi- EMDB-3206: Revisited cryo-EM structure of Inducible lysine decarboxylase com... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3206 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Revisited cryo-EM structure of Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | |||||||||
 Map data Map data | Reconstruction of E.coli Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | acid-stress / lysine decarboxylase / ATPase / RavA / LARA / cage | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysine decarboxylase / lysine catabolic process / lysine decarboxylase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances / guanosine tetraphosphate binding / ATP hydrolysis activity / ATP binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
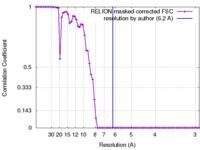
| Method | single particle reconstruction / cryo EM / Resolution: 6.2 Å | |||||||||
 Authors Authors | Kandiah E / Carriel D / Perard J / Malet H / Bacia M / Liu K / Chan SWS / Houry WA / Ollagnier de Choudens S / Elsen S / Gutsche I | |||||||||
 Citation Citation |  Journal: Elife / Year: 2014 Journal: Elife / Year: 2014Title: Assembly principles of a unique cage formed by hexameric and decameric E. coli proteins. Authors: Hélène Malet / Kaiyin Liu / Majida El Bakkouri / Sze Wah Samuel Chan / Gregory Effantin / Maria Bacia / Walid A Houry / Irina Gutsche /   Abstract: A 3.3 MDa macromolecular cage between two Escherichia coli proteins with seemingly incompatible symmetries-the hexameric AAA+ ATPase RavA and the decameric inducible lysine decarboxylase LdcI-is ...A 3.3 MDa macromolecular cage between two Escherichia coli proteins with seemingly incompatible symmetries-the hexameric AAA+ ATPase RavA and the decameric inducible lysine decarboxylase LdcI-is reconstructed by cryo-electron microscopy to 11 Å resolution. Combined with a 7.5 Å resolution reconstruction of the minimal complex between LdcI and the LdcI-binding domain of RavA, and the previously solved crystal structures of the individual components, this work enables to build a reliable pseudoatomic model of this unusual architecture and to identify conformational rearrangements and specific elements essential for complex formation. The design of the cage created via lateral interactions between five RavA rings is unique for the diverse AAA+ ATPase superfamily. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3206.map.gz emd_3206.map.gz | 28.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3206-v30.xml emd-3206-v30.xml emd-3206.xml emd-3206.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3206_fsc.xml emd_3206_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_3206.tif emd_3206.tif | 1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3206 http://ftp.pdbj.org/pub/emdb/structures/EMD-3206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3206 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3206 | HTTPS FTP |
-Validation report
| Summary document |  emd_3206_validation.pdf.gz emd_3206_validation.pdf.gz | 283.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3206_full_validation.pdf.gz emd_3206_full_validation.pdf.gz | 283 KB | Display | |
| Data in XML |  emd_3206_validation.xml.gz emd_3206_validation.xml.gz | 9.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3206 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3206 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3206 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3206 | HTTPS FTP |
-Related structure data
| Related structure data |  5fl2MC  3204C  3205C  5fkxC  5fkzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3206.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3206.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
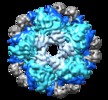
| Annotation | Reconstruction of E.coli Inducible lysine decarboxylase complexed with LARA domain of RavA ATPase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.46466 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex between a decamer of inducible lysine decarboxylase LdcI ...
| Entire | Name: Complex between a decamer of inducible lysine decarboxylase LdcI and ten LARA domains of the AAA+ ATPase RavA |
|---|---|
| Components |
|
-Supramolecule #1000: Complex between a decamer of inducible lysine decarboxylase LdcI ...
| Supramolecule | Name: Complex between a decamer of inducible lysine decarboxylase LdcI and ten LARA domains of the AAA+ ATPase RavA type: sample / ID: 1000 Oligomeric state: One homodecamer of LdcI bound to ten monomers of LARA domains of the AAA+ ATPase RavA Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 936 KDa / Theoretical: 936 KDa |
-Macromolecule #1: Inducible lysine decarboxylase
| Macromolecule | Name: Inducible lysine decarboxylase / type: protein_or_peptide / ID: 1 / Name.synonym: LdcI / Number of copies: 10 / Oligomeric state: Homodecamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 81.2 KDa / Theoretical: 81.2 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Inducible lysine decarboxylase / GO: lysine catabolic process / InterPro: Ornithine/lysine/arginine decarboxylase |
-Macromolecule #2: LARA domain of the AAA+ ATPase RavA
| Macromolecule | Name: LARA domain of the AAA+ ATPase RavA / type: protein_or_peptide / ID: 2 / Name.synonym: LARA domain of RavA / Number of copies: 10 / Oligomeric state: monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 12.405 KDa / Theoretical: 12.405 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: ATPase RavA / GO: ATP hydrolysis activity / InterPro: ATPase RavA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 6.5 Details: 50mM MES pH 6.5, 100mM NaCl, 0.2mM PLP, 1mM DTT, 0.01% glutaraldehyde |
| Grid | Details: Quantifoil 400 mesh 2/1 Glow discharged |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 91 K / Instrument: FEI VITROBOT MARK III / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 90 K / Max: 92 K / Average: 91 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification Legacy - Electron beam tilt params: 0 |
| Details | Automatic data acquisition with FEI EPU software |
| Date | Sep 14, 2012 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 911 / Average electron dose: 20 e/Å2 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 51660 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 51660 |
| Sample stage | Specimen holder: Nitrogen cooled / Specimen holder model: GATAN HELIUM |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Software | Name: Chimera, IMODFIT |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: cross-correlation |
| Output model |  PDB-5fl2: |
-Atomic model buiding 2
| Initial model | PDB ID: Chain - Chain ID: X |
|---|---|
| Software | Name: SITUS |
| Details | The residues 332-437 corresponding to the RavA LARA domain were fitted |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation |
| Output model |  PDB-5fl2: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)