[English] 日本語
 Yorodumi
Yorodumi- EMDB-31808: Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | |||||||||
 Map data Map data | postprocess_masked.mrc from Relion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cas12c / c2c3 / sgRNA / target DNA / CRISPR / RNA binding protein-RNA-DNA complex | |||||||||
| Biological species |  uncultured archaeon (environmental samples) / synthetic construct (others) uncultured archaeon (environmental samples) / synthetic construct (others) | |||||||||
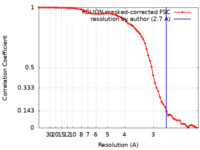
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Kurihara N / Hirano H / Tomita A / Kobayashi K | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structure of the type V-C CRISPR-Cas effector enzyme. Authors: Nina Kurihara / Ryoya Nakagawa / Hisato Hirano / Sae Okazaki / Atsuhiro Tomita / Kan Kobayashi / Tsukasa Kusakizako / Tomohiro Nishizawa / Keitaro Yamashita / David A Scott / Hiroshi ...Authors: Nina Kurihara / Ryoya Nakagawa / Hisato Hirano / Sae Okazaki / Atsuhiro Tomita / Kan Kobayashi / Tsukasa Kusakizako / Tomohiro Nishizawa / Keitaro Yamashita / David A Scott / Hiroshi Nishimasu / Osamu Nureki /    Abstract: RNA-guided CRISPR-Cas nucleases are widely used as versatile genome-engineering tools. Recent studies identified functionally divergent type V Cas12 family enzymes. Among them, Cas12c2 binds a CRISPR ...RNA-guided CRISPR-Cas nucleases are widely used as versatile genome-engineering tools. Recent studies identified functionally divergent type V Cas12 family enzymes. Among them, Cas12c2 binds a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA) and recognizes double-stranded DNA targets with a short TN PAM. Here, we report the cryo-electron microscopy structures of the Cas12c2-guide RNA binary complex and the Cas12c2-guide RNA-target DNA ternary complex. The structures revealed that the crRNA and tracrRNA form an unexpected X-junction architecture, and that Cas12c2 recognizes a single T nucleotide in the PAM through specific hydrogen-bonding interactions with two arginine residues. Furthermore, our biochemical analyses indicated that Cas12c2 processes its precursor crRNA to a mature crRNA using the RuvC catalytic site through a unique mechanism. Collectively, our findings improve the mechanistic understanding of diverse type V CRISPR-Cas effectors. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31808.map.gz emd_31808.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31808-v30.xml emd-31808-v30.xml emd-31808.xml emd-31808.xml | 22.7 KB 22.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31808_fsc.xml emd_31808_fsc.xml | 6.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_31808.png emd_31808.png | 116.2 KB | ||
| Masks |  emd_31808_msk_1.map emd_31808_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-31808.cif.gz emd-31808.cif.gz | 7.1 KB | ||
| Others |  emd_31808_additional_1.map.gz emd_31808_additional_1.map.gz emd_31808_half_map_1.map.gz emd_31808_half_map_1.map.gz emd_31808_half_map_2.map.gz emd_31808_half_map_2.map.gz | 23.5 MB 20.7 MB 20.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31808 http://ftp.pdbj.org/pub/emdb/structures/EMD-31808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31808 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31808 | HTTPS FTP |
-Validation report
| Summary document |  emd_31808_validation.pdf.gz emd_31808_validation.pdf.gz | 744.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31808_full_validation.pdf.gz emd_31808_full_validation.pdf.gz | 744.2 KB | Display | |
| Data in XML |  emd_31808_validation.xml.gz emd_31808_validation.xml.gz | 12.5 KB | Display | |
| Data in CIF |  emd_31808_validation.cif.gz emd_31808_validation.cif.gz | 17.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31808 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31808 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31808 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31808 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31808.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31808.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postprocess_masked.mrc from Relion | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08073 Å | ||||||||||||||||||||||||||||||||||||
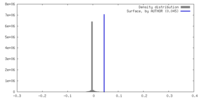
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_31808_msk_1.map emd_31808_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
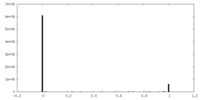
| Density Histograms |
-Additional map: calculated by deepEMhancer using run class001.mrc from Refine3D
| File | emd_31808_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | calculated by deepEMhancer using run_class001.mrc from Refine3D | ||||||||||||
| Projections & Slices |
| ||||||||||||
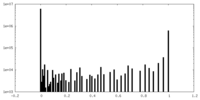
| Density Histograms |
-Half map: run half1 class001 unfil.mrc from Relion
| File | emd_31808_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | run_half1_class001_unfil.mrc from Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
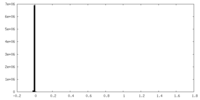
| Density Histograms |
-Half map: run half2 class001 unfil.mrc from Relion
| File | emd_31808_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | run_half2_class001_unfil.mrc from Relion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cas12c2-sgRNA-DNA complex
| Entire | Name: Cas12c2-sgRNA-DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Cas12c2-sgRNA-DNA complex
| Supramolecule | Name: Cas12c2-sgRNA-DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  uncultured archaeon (environmental samples) uncultured archaeon (environmental samples) |
-Supramolecule #2: Cas12c2
| Supramolecule | Name: Cas12c2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  uncultured archaeon (environmental samples) uncultured archaeon (environmental samples) |
-Supramolecule #3: sgRNA
| Supramolecule | Name: sgRNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Supramolecule #4: DNA
| Supramolecule | Name: DNA / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3-#4 |
|---|
-Macromolecule #1: Cas12c2
| Macromolecule | Name: Cas12c2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  uncultured archaeon (environmental samples) uncultured archaeon (environmental samples) |
| Molecular weight | Theoretical: 138.348938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIEEGKGHH HHHHMTKHSI PLHAFRNSGA DARKWKGRIA LLAKRGKETM RTLQFPLEMS EPEAAAINTT PFAVAYNAIE GTGKGTLFD YWAKLHLAGF RFFPSGGAAT IFRQQAVFED ASWNAAFCQQ SGKDWPWLVP SKLYERFTKA PREVAKKDGS K KSIEFTQE ...String: MKIEEGKGHH HHHHMTKHSI PLHAFRNSGA DARKWKGRIA LLAKRGKETM RTLQFPLEMS EPEAAAINTT PFAVAYNAIE GTGKGTLFD YWAKLHLAGF RFFPSGGAAT IFRQQAVFED ASWNAAFCQQ SGKDWPWLVP SKLYERFTKA PREVAKKDGS K KSIEFTQE NVANESHVSL VGASITDKTP EDQKEFFLKM AGALAEKFDS WKSANEDRIV AMKVIDEFLK SEGLHLPSLE NI AVKCSVE TKPDNATVAW HDAPMSGVQN LAIGVFATCA SRIDNIYDLN GGKLSKLIQE SATTPNVTAL SWLFGKGLEY FRT TDIDTI MQDFNIPASA KESIKPLVES AQAIPTMTVL GKKNYAPFRP NFGGKIDSWI ANYASRLMLL NDILEQIEPG FELP QALLD NETLMSGIDM TGDELKELIE AVYAWVDAAK QGLATLLGRG GNVDDAVQTF EQFSAMMDTL NGTLNTISAR YVRAV EMAG KDEARLEKLI ECKFDIPKWC KSVPKLVGIS GGLPKVEEEI KVMNAAFKDV RARMFVRFEE IAAYVASKGA GMDVYD ALE KRELEQIKKL KSAVPERAHI QAYRAVLHRI GRAVQNCSEK TKQLFSSKVI EMGVFKNPSH LNNFIFNQKG AIYRSPF DR SRHAPYQLHA DKLLKNDWLE LLAEISATLM ASESTEQMED ALRLERTRLQ LQLSGLPDWE YPASLAKPDI EVEIQTAL K MQLAKDTVTS DVLQRAFNLY SSVLSGLTFK LLRRSFSLKM RFSVADTTQL IYVPKVCDWA IPKQYLQAEG EIGIAARVV TESSPAKMVT EVEMKEPKAL GHFMQQAPHD WYFDASLGGT QVAGRIVEKG KEVGKERKLV GYRMRGNSAY KTVLDKSLVG NTELSQCSM IIEIPYTQTV DADFRAQVQA GLPKVSINLP VKETITASNK DEQMLFDRFV AIDLGERGLG YAVFDAKTLE L QESGHRPI KAITNLLNRT HHYEQRPNQR QKFQAKFNVN LSELRENTVG DVCHQINRIC AYYNAFPVLE YMVPDRLDKQ LK SVYESVT NRYIWSSTDA HKSARVQFWL GGETWEHPYL KSAKDKKPLV LSPGRGASGK GTSQTCSCCG RNPFDLIKDM KPR AKIAVV DGKAKLENSE LKLFERNLES KDDMLARRHR NERAGMEQPL TPGNYTVDEI KALLRANLRR APKNRRTKDT TVSE YHCVF SDCGKTMHAD ENAAVNIGGK FIADIEK |
-Macromolecule #2: sgRNA
| Macromolecule | Name: sgRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  uncultured archaeon (environmental samples) uncultured archaeon (environmental samples) |
| Molecular weight | Theoretical: 36.147414 KDa |
| Sequence | String: GGAUACCACC CGUGCAUUUC UGGAUCAAUG AUCCGUACCU CAAUGUCCGG GCGCGCAGCU AGAGCGACCU GAAGAAAUUC AGGUUGGGU UUGAGGGGAA AUUAGGUGCG CUU |
-Macromolecule #3: target DNA (target strand)
| Macromolecule | Name: target DNA (target strand) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.080494 KDa |
| Sequence | String: (DA)(DG)(DG)(DT)(DT)(DG)(DC)(DC)(DA)(DA) (DG)(DC)(DG)(DC)(DA)(DC)(DC)(DT)(DA)(DA) (DT)(DT)(DT)(DC)(DC)(DC)(DA)(DT)(DT) (DT)(DT)(DA)(DG) |
-Macromolecule #4: target DNA (non target strand)
| Macromolecule | Name: target DNA (non target strand) / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 10.218596 KDa |
| Sequence | String: (DC)(DT)(DA)(DA)(DA)(DA)(DT)(DG)(DG)(DG) (DA)(DA)(DA)(DT)(DT)(DA)(DG)(DG)(DT)(DG) (DC)(DG)(DC)(DT)(DT)(DG)(DG)(DC)(DA) (DA)(DC)(DC)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average exposure time: 2.6 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7v94: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)